Abstract
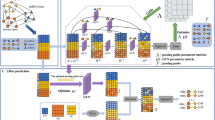
An increasing number of studies and experiments have demonstrated that long noncoding RNAs (lncRNAs) have a massive impact on various biological processes. Predicting potential associations between lncRNAs and diseases not only can improve our understanding of the molecular mechanisms of human diseases but also can facilitate the identification of biomarkers for disease diagnosis, treatment, and prevention. However, identifying such associations through experiments is costly and demanding, thereby prompting researchers to develop computational methods to complement these experiments. In this paper, we constructed a novel model called RWSF-BLP (a novel lncRNA-disease association prediction model using Random Walk-based multi-Similarity Fusion and Bidirectional Label Propagation), which applies an efficient random walk-based multi-similarity fusion (RWSF) method to fuse different similarity matrices and utilizes bidirectional label propagation to predict potential lncRNA-disease associations. Leave-one-out cross-validation (LOOCV) and 5-fold cross-validation (5-fold-CV) were implemented in the evaluation RWSF-BLP performance. Results showed that, RWSF-BLP has reliable AUCs of 0.9086 and 0.9115 ± 0.0044 under the framework of LOOCV and 5-fold-CV and outperformed other four canonical methods. Case studies on lung cancer and leukemia demonstrated that potential lncRNA-disease associations can be predicted through our method. Therefore, our method can accurately infer potential lncRNA-disease associations and may be a good choice in future biomedical research.








Similar content being viewed by others
References
Adomavicius G, Tuzhilin A (2005) Toward the next generation of recommender systems: a survey of the state-of-the-art and possible extensions. IEEE Trans Knowl Data Eng 17:734–749
Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS (2010) lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res 39:D146–D151
Bao Z, Yang Z, Huang Z, Zhou Y, Cui Q, Dong D (2019) LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res 47:D1034–D1037
Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M (2007) Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 123:215–229
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q (2012) LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 41:D983–D986
Chen X, Yan G-Y (2013) Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics 29:2617–2624
Chen X (2016) KATZLDA: KATZ measure for the lncRNA-disease association prediction. Sci Rep 5:16840
Chen Shifeng W, Hong LN, Hongling W, Yushu W, Qianqing T, Huiyuan S, Chengming S (2016) BMC genomics. Biomed Pharmacother 82:583–588
Chen X (2015) Predicting lncRNA-disease associations and constructing lncRNA functional similarity network based on the information of miRNA. Sci Rep 5:13186
Chen X, Yan CC, Zhang X, You Z-H (2017) Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform 18:558–576
Chen X, Yang J-R, Guan N-N, Li J-Q (2018) GRMDA: graph regression for MiRNA-disease association prediction. Front Physiol 9:92
Chen Q, Lai D, Lan W, Wu X, Chen B, Chen YP, Wang J (2019) ILDMSF: inferring associations between long non-coding RNA and disease based on multi-similarity fusion. IEEE/ACM Trans Comput Biol Bioinform. https://doi.org/10.1109/TCBB.2019.2936476
Congrains A, Kamide K, Oguro R, Yasuda O, Miyata K, Yamamoto E, Kawai T, Kusunoki H, Yamamoto H, Takeya Y, Yamamoto K (2012) Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 220:449–455
Cui D, Yu C-H, Liu M, Xia Q-Q, Zhang Y-F, Jiang W-L (2016) Long non-coding RNA PVT1 as a novel biomarker for diagnosis and prognosis of non-small cell lung cancer. Tumor Biol 37:4127–4134
Cui T, Zhang L, Huang Y, Yi Y, Tan P, Zhao Y, Hu Y, Xu L, Li E, Wang D (2018) MNDR v2.0: an updated resource of ncRNA-disease associations in mammals. Nucleic Acids Res 46:D371–D374
Detterbeck FC, Boffa DJ, Kim AW (2017) The eighth edition lung cancer stage classification. Chest 151:193–203
Dieter C, Lourenco ED, Lemos NE (2020) Association of long non-coding RNA and leukemia: a systematic review. Gene 735:144405
Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS (2008) NRED: a database of long noncoding RNA expression. Nucleic Acids Res 37:D122–D126
Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12:861
Lan W, Li M, Zhao K, Liu J, Wu F-X, Pan Y, Wang J (2016) LDAP: a web server for lncRNA-disease association prediction. Bioinformatics 33:458–460
Fu G, Wang J, Domeniconi C, Yu G (2018) Matrix factorization-based data fusion for the prediction of lncRNA-disease associations. Bioinformatics 34:1529–1537
Ferrando AA, López-Otín C (2019) Clonal evolution in leukemia. Nucleic Acids Res 47:D1034–D1037
Gao S, Zhou B, Li H, Huang X, Wu Y, Xing C, Yu X, Ji Y (2018) Long noncoding RNA HOTAIR promotes the self-renewal of leukemia stem cells through epigenetic silencing of p15. Exp Hematol 67:32–40
Garitano-Trojaola A (2018) Deregulation of linc-PINT in acute lymphoblastic leukemia is implicated in abnormal proliferation of leukemic cells. Oncotarget 9:12842
Greenlee RT, Murray T, Bolden S, Wingo PA (2000) Cancer statistics. CA Cancer J Clin 50:7–33
Guan N-N, Wang C-C, Zhang L, Huang L, Li J-Q, Piao X (2020) In silico prediction of potential miRNA-disease association using an integrative bioinformatics approach based on kernel fusion. J Cell Mol Med 24:573–587
Guo ZH, You ZH, Wang YB, Yi HC, Chen ZH (2019) A learning-based method for lncRNA-disease association identification combing similarity information and rotation forest. iScience 19:786–795
Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES (2013) Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154:240–251
Huang Y, Yuan K, Tang M, Yue J, Bao L, Wu S, Zhang Y, Li Y, Wang Y, Ou X et al (2020) Melatonin inhibiting the survival of human gastric cancer cells under ER stress involving autophagy and Ras-Raf-MAPK signalling. J Cell Mol Med. https://doi.org/10.1111/jcmm.16237
Huang Y-A, Chen X, You Z-H, Huang D-S, Chan KCC (2016a) ILNCSIM: improved lncRNA functional similarity calculation model. Oncotarget 7:25902
Huang C, Liu S, Wang H, Zhang Z, Yang Q, Gao F (2016b) LncRNA PVT1 overexpression is a poor prognostic biomarker and regulates migration and invasion in small cell lung cancer. Am J Transl Res 8:5025
Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom 8:39
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61:69–90
Lan W, Li M, Zhao K, Liu J, Wu F-X, Pan Y, Wang J (2016) LDAP: a web server for lncRNA-disease association prediction. Bioinformatics 33:458–460
Lan W, Lai D, Chen Q, Wu X, Chen B, Liu J, Wang J, Chen Y-PP (2020) LDICDL: LncRNA-disease association identification based on collaborative deep learning. IEEE/ACM Trans Comput Biol Bioinform 14:1–1
Lei L, Xia S, Liu D, Li X, Feng J, Zhu Y, Hu J, Xia L, Guo L, Chen F et al (2018) Genome-wide characterization of lncRNAs in acute myeloid leukemia. Brief Bioinform 19:627–635
Li W, Huang K, Wen F, Cui G, Guo H, Zhao S (2017) Genetic variation of lncRNA GAS5 contributes to the development of lung cancer. Oncotarget 8:91025
Li Y, Li J, Bian N (2019) DNILMF-LDA: prediction of lncRNA-disease associations by dual-network integrated logistic matrix factorization and Bayesian optimization. Genes 10:608
Liu H, Ren G, Chen H, Liu Q, Yang Y, Zhao Q (2020) Predicting lncRNA-miRNA interactions based on logistic matrix factorization with neighborhood regularized. Knowl Based Syst 8191:105261
Liu J, Lee W, Jiang Z, Chen Z, Jhunjhunwala S, Haverty PM, Gnad F, Guan Y, Gilbert HN, Stinson J (2012) Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res 22:2315–2327
Lu C, Yang M, Luo F, Wu F-X, Li M, Pan Y, Li Y, Wang J (2018) Prediction of lncRNA-disease associations based on inductive matrix completion. Bioinformatics 34:3357–3364
Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F (2000) Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 5:211–220
Omer A, Singh P, Yadav NK, Singh RK (2015) MicroRNAs: role in leukemia and their computational perspective. Wiley Interdiscip Rev RNA 6:65–78
Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N, Lilja P, Lukk M et al (2006) ArrayExpress-a public database of microarray experiments and gene expression profiles. Nucleic Acids Res f35:D747–D750
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136:629–641
Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E et al (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129:1311–1323
Saito T, Rehmsmeier M (2015) The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PloS One 10:e0118432
Spiess PE, Dhillon J, Baumgarten AS, Johnstone PA (2016) Pathophysiological basis of human papillomavirus in penile cancer: key to prevention and delivery of more effective therapies. CA Cancer J Clin 66:481–495
Sumathipala M, Maiorino E, Weiss ST, Sharma A (2019) Network diffusion approach to predict lncRNA disease associations using multi-type biological networks: LION. Front Physiol 10:888
Sun J, Shi H, Wang Z, Zhang C, Liu L, Wang L, He W, Hao D, Liu S, Zhou M (2014) Inferring novel lncRNA-disease associations based on a random walk model of a lncRNA functional similarity network. Mol Biosyst 10:2074–2081
Zhao T, Xu J, Liu L, Bai J, Xu C, Xiao Y, Li X, Zhang L (2014) Identification of cancer-related lncRNAs through integrating genome regulome and transcriptome features. Mol BioSyst 11:126–136
van Laarhoven T, Nabuurs SB, Marchiori E (2011) Gaussian interaction profile kernels for predicting drug-target interaction. Bioinformatics 10:3036–3043
Verhaegh GW, Verkleij L, Vermeulen SHHM, den Heijer M, Witjes JA, Kiemeney LA (2008) Polymorphisms in the H19 gene and the risk of bladder cancer. Eur Urol 54:1118–1126
Wang D, Wang J, Lu M, Song F, Cui Q (2010) Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics 26:1644–1650
Wang L, Wang Y, Li H, Feng X, Yuan D, Yang J (2019) A bidirectional label propagation based computational model for potential microbe-disease association prediction. Front Microbiol 27:684
Wang Y, Yu G, Wang J, Fu G, Guo M, Domeniconi C (2020) Weighted matrix factorization on multi-relational data for LncRNA-disease association prediction. Methods 173:32–43
Wen F, Cao Y-X, Luo Z-Y, Liao P, Lu Z-W (2018) LncRNA MALAT1 promotes cell proliferation and imatinib resistance by sponging miR-328 in chronic myelogenous leukemia. Biochem Biophysl Res Commun 507:1–8
White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA (2014) Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol 15:429
Xie G, Huang S, Luo Yu, Ma L, Lin Z, Sun Y (2019) LLCLPLDA: a novel model for predicting lncRNA-disease associations. Mol Genet Genom 294:1477–1486
Yao H, Duan M, Lin L, Wu C, Fu X, Wang H, Guo L, Chen W, Huang L, Liu D et al (2017) TET2 and MEG3 promoter methylation is associated with acute myeloid leukemia in a Hainan population. Oncotarget 8:18337
Yao D, Zhan X, Zhan X, Kwoh CK, Li P, Wang J (2020) A random forest based computational model for predicting novel lncRNA-disease associations. BMC Bioinform 21:1–18
Yang D, Zhang X, Zhang X, Xu Y (2017) The progress and current status of immunotherapy in acute myeloid leukemia. Ann Hematol 96:1965–1982
Yu F, Zheng J, Mao Y, Dong P, Li G, Lu Z, Guo C, Liu Z, Fan X (2015) Long non-coding RNA APTR promotes the activation of hepatic stellate cells and the progression of liver fibrosis. Biochem Biophys Res Commun 463:679–685
Yu J, Xuan Z, Feng X, Zou Q, Wang L (2019) A novel collaborative filtering model for LncRNA-disease association prediction based on the Native Bayesian classifier. BMC Bioinform 20:396
Zeng M, Lu C, Zhang F, Li Y, Wu F-X, Li Y, Li M (2020) SDLDA: lncRNA-disease association prediction based on singular value decomposition and deep learning. Methods 179:73–80
Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A (2003) A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metabol 88:5119–5126
Zhang H, Liang Y, Peng C, Han S, Du W, Li Y (2019) Long noncoding RNA and protein interactions: from experimental results to computational models based on network methods. Math Biosci 20:1284
Zhang Y, Chen M, Li A, Cheng X, Jin H, Liu Y (2020a) LDAI-ISPS: LncRNA-disease associations inference based on integrated space projection scores. Int J Mol Sci 21:4
Zhang Y, Ye F, Xiong D, Gao X (2020b) LDNFSGB: prediction of long non-coding rna and disease association using network feature similarity and gradient boosting. BMC Bioinform 21:1–27
Zhu Z, Song L, He J, Sun Y, Liu X, Zou X (2015) Ectopic expressed long non-coding RNA H19 contributes to malignant cell behavior of ovarian cancer. BMC Genom 8:10082
Funding
This work was supported by the National Natural Science Foundation of China (62002070, 618002072), the Natural Science Foundation of Guangdong Province (2018A030313389), the Science and Technology Plan Project of Guangdong Province (2019B010139002, 2018B030323026, 2017A040405050, 2016B030306004), and the Science and Technology Program of Guangzhou (201902020006, 201902020012, 201907010021).Yuping Sun is the corresponding author.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Stefan Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xie, G., Huang, B., Sun, Y. et al. RWSF-BLP: a novel lncRNA-disease association prediction model using random walk-based multi-similarity fusion and bidirectional label propagation. Mol Genet Genomics 296, 473–483 (2021). https://doi.org/10.1007/s00438-021-01764-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-021-01764-3