Abstract
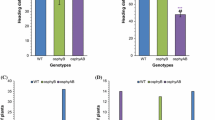
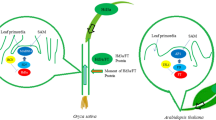
Heading date is one of most important agronomic traits in rice. Flowering regulatory mechanisms have been elucidated in many cultivars through various approaches. Although study about flowering has been extensively examined in rice, but contributions of floral regulators had been poorly understood in a common genetic background for rice grown under paddy conditions. Thus, we compared the expression of 10 flowering-time genes — OsMADS50, OsMADS51, OsVIL2, OsPhyA, OsPhyB, OsPhyC, Ghd7, Hd1, OsGI, and OsTrx1 — in the same genetic background for ‘Dongjin’ rice (Oryza sativa) grown under paddy conditions when days were longer than 13.5 h. Whereas the wild type (WT) rice flowered 105 days after sowing, the latest mutant to do so was ostrx1, flowering 53 d later. This indicated that the gene is the strongest inducer among all of those examined. Mutations in OsMADS50 delayed flowering by 45 d when compared with the WT, suggesting that this MADS gene is another strong positive element. The third positive element was OsVIL2; mutations in the gene caused plants to flower 27 d late. In contrast, the double phytochrome mutant osphyA osphyB flowered 44 d earlier than the WT. The single mutant osphyB and the double mutant osphyB osphyC did the same, although not as early as the osphyA osphyB double mutant. These results demonstrated that phytochromes are major inhibitors under paddy conditions. Mutations in Ghd7 accelerated flowering by 34 d, indicating that the gene is also a major inhibitor. The hd1 mutants flowered 16 d earlier than the WT while a mutation in OsGI hastened flowering by 10 d, suggesting that both are weak flowering repressors. Of the two florigen genes (Hd3a being the other one), RFT1 played a major role under paddy conditions. Its expression was strongly promoted by Ehd1, which was negatively controlled by Ghd7. Here we show that phytochromes strongly inhibit flowering and OsTrx1 and OsMADS50 significantly induce flowering under paddy conditions through Ghd7-Ehd1-RFT1 pathway. Thus, we may be able to control heading date under paddy conditions through manipulating those genes, Ghd7, Ehd1 and RFT1.
Similar content being viewed by others
References
Araki T, Komeda Y (1993) Flowering in darkness in Arabidopsis thaliana. Plant J 4:801–811
Choi SC, Lee S, Kim SR, Lee YS, Liu C, Cao X, An G (2014) Trithorax group protein OsTrx1 controls flowering time in rice via interaction with Ehd3. Plant Physiol 164:1326–1337
Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FTlike gene expression independently of Hd1. Genes Dev 18:926–936
Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999). GIGANTEA: A circadian clockcontrolled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane spanning domains. EMBO J 18:4679–4688
Gomez-Ariza J, Galbiati F, Gorett D, Brambilla V, Shrestha R, Pappolla A, Courtois B, Fornara F (2015) Loss of floral repressor function adapts rice to higher latitudes in Europe. J Exp Bot 66:2027–2039
Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, An K, Han MJ, Sung RJ, Choi HS, Yu JH, Choi JH, Cho SY, Cha SS, Kim SI, An G (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22:561–570
Jeong DH, An S, Kang HG, Moon S, Han JJ, Park S, Lee HS, An K, An G (2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol 130:1636–1644
Jeong DH, Lee S, Kim SL, Hwang I, An G (2007) Regulation of brassinosteroid responses by phytochrome B in rice. Plant Cell Environ 30:590–599
Jeong HJ, Yang J, Yi J, An G (2015) Controlling flowering time by histone methylation and acetylation in Arabidopsis and rice. J Plant Biol 58:203–210
Kim SL, Lee S, Kim HJ, Nam HG, An G (2007) OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol 145:1484–1494
Kim SL, Choi M, Jung KH, An G (2013) Analysis of the earlyflowering mechanisms and generation of T-DNA tagging lines in Kitaake, a model rice cultivar. J Exp Bot 64:4169–4182
Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43:1096–1105
Komiya R, Yokoi S, Shimamoto K (2009) A gene network for longday flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136:3443–3450
Koo BH, Yoo SC, Park JW, Kwon CT, Lee BD, An G, Zhang Z, Li J, Li Z, Paek NC (2013) Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol Plant 6:1877–1888
Lee S, Kim J, Han JJ, Han MJ, An G (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J 38:754–764
Lee YS, An G (2015a) OsGI controls flowering time by modulating rhythmic flowering time regulators preferentially under short days in rice. J Plant Biol 58:137–145
Lee YS, An G (2015b) Regulation of flowering time in rice. J Plant Biol 58:353–360
Lee YS, Jeong DH, Lee DY, Yi J, Ryu CH, Kim SL, Jeong HJ, Choi SC, Jin P, Yang J, Cho LH, Choi H, An G (2010) OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J 63:18–30
Lee YS, Lee DY, Cho LH, An G (2014) Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice 7:31
Lin HX, Yamamoto T, Sasaki T, Yano M (2000) Characterization and detection of epistatic interactions of three QTLs, Hd1, Hd2 and Hd3, controlling heading date in rice using nearly isogenic lines. Theor Appl Genet 101:1021–1028
Matsubara K, Yamanouchi U, Wang ZX, Minobe Y, Izawa T, Yano M (2008) Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol 148:1425–1435
Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, Coupland G (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17:2255–2270
Ogiso-Tanaka E, Matsubara K, Yamamoto S-i, Nonoue Y, Wu J, Fujisawa H, Ishikubo H, Tanaka T, Ando T, Matsumoto T, Yano M (2013) Natural variation of the RICE FLOWERING LOCUS T 1 contributes to flowering time divergence in rice. PLoS One 8:e75959
Osugi A, Itoh H, Ikeda-Kawakatsu K, Takano M, Izawa T (2011) Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol 157:1128–1137
Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285:1579–1582
Park SJ, Kim SL, Lee S, Je BI, Piao HL, Park SH, Kim CM, Ryu CH, Park SH, Xuan YH, Colasanti J, An G, Han CD (2008) Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J 56:1018–1029
Ryu CH, Lee S, Kim SL, Lee YS, Choi SC, Jeong HJ, Yi J, Park SJ, Han CD, An G (2009) OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ 32:1412–1427
Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288:1613–1616
Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio BA, Nagamura Y (2013) RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res 41:D1206–1213
Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, Ishizuka T, Yano M, Nishimura M, Miyao A, Hirochika H, Shinomura T (2005) Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17:3311–3325
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316:1033–1036
Uga Y, Nonoue Y, Liang ZW, Lin HX, Yamamoto S, Yamanouchi U, Yano M (2007) Accumulation of additive effects generates a strong photoperiod sensitivity in the extremely late-heading rice cultivar ‘Nona Bokra’. Theor Appl Genet 114:1457–1466
Wu CY, You CJ, Li CS, Long T, Chen GX, Byrne ME, Zhang QF (2008) RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc Natl Acad Sci USA 105:12915–12920
Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40:761–767
Yang J, Lee S, Hang R, Kim SR, Lee YS, Cao X, Amasino R, An G (2013) OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J 73:566–578
Yang L, Liu T, Li B, Sui Y, Chen J, Shi J, Wing RA, Chen M (2012) Comparative sequence analysis of the Ghd7 orthologous regions revealed movement of Ghd7 in the grass genome. PLoS ONE 7:e50236
Yano M, Sasaki T (1997) Genetic and molecular dissection of quantitative traits in rice. Plant Mol Biol 35:145–153
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2483
Yi J, An G (2013) Utilization of T-DNA tagging lines in rice. J Plant Biol 56:85–90
Yu SB, Li JX, Xu CG, Tan YF, Li XH, Zhang Q (2002) Identification of quantitative trait loci and epistatic interactions for plant height and heading date in rice. Theor Appl Genet 104:619–625
Zhao J, Chen H, Ren D, Tang H, Qiu R, Feng J, Long Y, Niu B, Chen D, Zhong T, Liu YG, Guo J (2015) Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/ RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). New Phytol 208:936–948
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Lee, YS., Yi, J., Jung, KH. et al. Comparison of rice flowering-time genes under paddy conditions. J. Plant Biol. 59, 238–246 (2016). https://doi.org/10.1007/s12374-016-0029-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-016-0029-0