Abstract
Beta thalassemia major is a common genetic disorder characterized by the reduced production or absence of beta globin, a product of the haemoglobin subunit beta (HBB) gene. Every year, approximately 10,000–12,000 children with thalassemia major are born in India. Molecular methodologies like ARMS (amplification-refractory mutation system)-PCR (polymerase chain reaction) and capillary sequencing are used to detect HBB gene mutations. It is, however, challenging to achieve comprehensive coverage of the HBB gene by these methods. Next-generation sequencing (NGS) can be used to circumvent these problems. Commercial NGS panels are prohibitively expensive and hence are not routinely implemented in most laboratories. We have developed a cost-effective, highly sensitive and specific, indigenous targeted NGS assay for detecting mutations in the HBB gene. Using this custom NGS assay, we processed 1530 samples (3017 alleles), in which we detected a spectrum of 48 pathogenic/likely pathogenic variants (mutations); IVS-I-5 (c.92+5G>C) was the most common mutation detected (allele frequency (AF) 44.55%), followed by the 619-bp deletion (AF 10.74%), c.92+1G>T (AF 6.99%), c.27_28insG (AF 6.23%), c.47G>A (AF 5.77%) and c.126_129delCTTT (AF 4.71%). Additionally, we discovered a novel mutation (c.7delC) that was submitted to the HbVar database (HbVar ID 3193) as a variant of unknown significance (VUS), probably pathogenic. The targeted NGS assay developed during this study was validated using orthogonal methods and showed excellent correlation with currently available molecular methods. Additionally, this targeted NGS assay was used to analyse the mutation spectrum of the largest beta thalassemia cohort from India.
Similar content being viewed by others
Introduction
Beta thalassemia major is one of the most common genetic disorders in India with reported carrier frequencies between 3 and 18% [1]. The carrier rate for beta thalassemia varies from 1 to 3% in the southern parts of the country and from 3 to 15% in the northern parts of the country. Expanded carrier screening and population-based screening methods have been employed to identify couples whose children might be at risk of being affected with beta thalassemia major. Despite these initiatives, beta thalassemia major continues to be a significant health problem, causing considerable mortality and morbidity. The disorder is caused owing to the presence of biallelic pathogenic/likely pathogenic variants (this term is interchangeably used with the term ‘mutations’ in this manuscript) in the haemoglobin subunit beta (HBB) gene [2]. Numerous studies have described the mutation spectrum observed in different geographic regions of India [3]. The most commonly used molecular techniques used in these studies include end-point PCR, reverse dot blot analysis [4], amplification-refractory mutation system polymerase chain reaction (ARMS PCR) [5] and Sanger/capillary sequencing (CE-Seq) of the entire gene [6]. ARMS PCR is mainly used to screen the commonest mutations prevalent in a particular geographical area; if this initial screening is negative, complete HBB gene sequencing is performed. With the gradual advancement of molecular technologies, CE-Seq has become the preferred method for the detection of pathogenic variants in the HBB gene. CE-Seq technology, however, has its own disadvantages like being labour intensive, requiring extensive manual analysis and difficulty in differentiating true small peaks from background noise. Next-generation sequencing (NGS) can be utilized to analyse a number of genes implicated in a particular phenotype simultaneously, a process that would otherwise require extensive resources, time and cost by CE-Seq. NGS approaches, however, have their own limitations. Commercially available panels are expensive, end up using excessive sequencing space and generally produce unnecessary information. We have developed and validated a highly multiplexed NGS-based low-cost custom assay for the HBB gene, which can simultaneously detect small sequence variants in both the exonic and intronic regions as well the commonly encountered 619-bp deletion (NG_000007.3:g.71609_72227del619) in the HBB gene. In addition, here, we discuss the analysis of a cohort of 1530 samples, which, to the best of our knowledge, is the largest molecular analysis of HBB gene mutations from India.
Materials and methods
Sample collection and DNA extraction
All samples included in this study were of individuals clinically diagnosed with thalassemia major and other haemoglobinopathies or carriers of these conditions. Clinical suspicion was augmented with HPLC/Hb electrophoresis prior to molecular analysis. HBB gene sequencing was requested for these samples to aid patient management. Samples were stored at 4 °C until DNA extraction. Two millilitres of peripheral blood was collected in K2-EDTA vacutainers from patients. Genomic DNA was extracted from 150 μL of peripheral blood and eluted in 200 μL of elution buffer using the DNeasy Blood and Tissue Kit (Qiagen, Germany), per the manufacturer’s instructions.
Long-range PCR
The entire HBB gene region encompassing the 5′ untranslated region (UTR), all 3 exons, both the introns and the 3′ UTR and untranslated regions of the gene was amplified as a single 2.3-kb amplicon. PCR conditions and primers were adapted from Wang et al. [7]. Briefly, the PCR was performed in a 20-μL reaction mix containing 200 nM of each primer (βF, 5-ACGGCTGTCATCACTTAGAC-3, GenBank HUMHBB sequence nucleotides 62010–62029; βR2, 5-CAGATTCCGGGTCACTGTG-3, sequence nucleotides 64299–64281; genomic coordinates of the amplicon are 11:5227179-5224926), final 1x HotStar buffer, HotStar Taq polymerase enzyme (cat. no. 203203; Qiagen, Germany), 2 mM MgCl2, and 200 nM dNTPs. The thermal cycling conditions were as follows: initial hold at 95 °C for 15 min; 25 cycles consisting of 95 °C for 20 s, 61 °C for 30 s, 72 °C for 75 s, and final extension at 72 °C for 5 min.
NGS library preparation and sequencing
The long-range PCR products of the HBB gene for each sample were purified using the Purelink PCR Purification Kit (Invitrogen, CA, USA). The amplicons were quantified using the Qubit system (Life Technologies, CA, USA) with the Broad range Qubit reagent (Thermo Fischer Scientific; catalogue no. Q32853). Tagmentation was performed using the Nextera XT library preparation kit (Illumina, CA, USA). The multiplex PCR master mix supplied with the kit was used for the subsequent indexing PCR. Custom indexing primers compatible with the Illumina sequencing platform were used at a final concentration of 200 nM. Cycling conditions comprised initial denaturation at 95 °C for 15 min followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 63 °C for 45 s, and extension at 72 °C for 90 s. The ramp rate was reduced to 1.5 °C/s during the cycling steps. This was followed by a final hold step for extension at 72 °C for 10 min. The index PCR products were quantified using the Qubit system. After this, the indexed PCR products were normalized in terms of molarity and subsequently pooled to form a single library. The pooled library was purified using the Purelink PCR Purification Kit (cat no. K310001; Thermo Fisher Scientific, MA, USA) and diluted to a final concentration of 4 nM using resuspension buffer (RSB; Illumina, CA, USA). The library was denatured for 5 min using 0.2 N NaOH and neutralized using HT1 buffer (Illumina, CA, USA). The HBB sequencing libraries were typically pooled at this point with other NGS libraries and further diluted to a final concentration of 14 pM, 9 pM, or 1.3 pM, respectively, depending on whether the MiSeq v3, MiSeq v2, or the NextSeq MidOutput sequencing kits were used. The pooled libraries included a 5% phiX library spike (Illumina, CA, USA) as a control and diversity enhancer. The samples were loaded onto an Illumina MiSeq/NextSeq cartridge (Illumina, CA, USA) and sequenced in 2*150 or 2*250 modes using Illumina’s sequencing by synthesis (SBS) chemistry.
Real-time ARMS PCR for detecting the c.92+5(G>C) variant (conventional IVS-I-5 mutation)
A real-time ARMS PCR was performed for 56 samples using in-house designed primers. The ARMS primers exploit the use of an additional extra mismatch on the 2nd to the last base at the 3′ end of the oligonucleotide primers to increase stringency of the amplification [8]. Briefly, the PCR was performed in a 10-μL reaction mix containing 200 nM in-house designed ARMS primers, 0.25× SYBR, 1× HotStar buffer, Hotstar Taq polymerase enzyme (cat. no. 203203; Qiagen, Germany), 2 mM MgCl2 and 200 nM dNTPs. The thermal cycling conditions were as follows: initial hold at 95 °C for 15 min; 35 cycles consisting of 95 °C for 10 s and 61 °C for 25 s (with data acquisition on green channel) followed by melt curve analysis.
End-point PCR (PCR followed by gel electrophoresis)
To confirm the large 619-bp deletion, the PCR products were electrophoresed on a 0.8% agarose gel to visually confirm the deletion. As mentioned earlier, the expected band size of the normal long-range PCR product is 2.3 kb. The size of the PCR products in the samples harbouring the 619-bp deletion is approximately 1.7 kb. End-point PCR was performed for a total of 40 samples. The PCR products were visualized on the BioRad gel documentation system.
CE-seq
Capillary sequencing was performed using 10 ng of the long-range PCR product as the template with primers and conditions adapted from Chan et al. [9]. Briefly, cycle sequencing was performed in a 10-μL reaction mix containing the 5× sequencing buffer, BigDye Terminator v3.1 Ready Reaction mix, 1 M betaine, 10 ng DNA, and 350 nM of the sequencing primer using standard thermal cycling conditions as prescribed by the manufacturer (Applied Biosystems). The cycle-sequencing products were purified using the ethanol/sodium acetate/EDTA method as described in Applied Biosystems’ BigDye Terminator v3.1 cycle sequencing kit protocol (part no. 4337035 Rev A; 09/2002). CE-seq was performed for a total of 40 samples on an ABI 3500 genetic analyser.
Bioinformatic analysis
For capillary sequencing, the files generated on the ABI 3500 Genetic Analyser were analysed manually using 4 peaks (Nucleobytes, The Netherlands) and with Mutation Surveyor v5.0 (Softgenetics, PA, USA) and SeqScape v3.0 (Life Technologies, CA, USA). NGS data were analysed using an in-house developed bioinformatics pipeline. Alignment to the reference genome (GRCH37/hg19 version) was performed using the Burrows Wheeler Aligner (BWA), and the resulting BAM files were manually analysed for variants using the GATK Variant Caller (BaseSpace BWA Enrichment Workflow v2.1.1. with BWA 0.7.7-isis-1.0.0 and GATK v1.6-23-gf0210b3). Visualization of BAM files was performed using GenomeBrowse v2.1.1 (Golden Helix, MT, USA). For identification of the 619-bp deletion, the presence of the deletion in the data can be unambiguously determined. A sharp cliff can be seen on either side of the deletion, and the reads on either side of the cliff have a readily discernible difference. As the sequencing depth is > 500× on average, identification of the 619-bp deletion is unambiguous and easy to identify, in both, the heterozygous and homozygous states (supplementary figure S1). A significant drop in coverage may indicate the presence of other large deletions; however, this would need to be confirmed with an orthogonal method.
Results
Demographics
Informed consent was obtained from all subjects. Of the 1530 patients tested, 896 (58.56%) were males and 634 (41.44%) were females (Supplementary Table S1). There were 1144 (74.77%) patients up to 18 years of age and 386 (25.23%) patients above 18 years of age tested in our study.
NGS
A total of 1530 samples were analysed for the presence of pathogenic/likely pathogenic mutations in the HBB gene at GenePath Dx between December 2015 and April 2019. All 1530 samples tested positive for either a biallelic pathogenic variant (homozygous or compound heterozygous) or a heterozygous pathogenic variant in the HBB gene, which implies consistency between the phenotypes and genotypes of all the samples tested in the study (diagnostic yield = 100%).
Eight hundred and forty-seven individuals (55.36%) harboured a single mutation in the homozygous/hemizygous state whereas 640 individuals (41.83%) harboured compound heterozygous mutations. Forty-three individuals (2.81%) were carriers for thalassemia and other haemoglobinopathies. The clinical diagnoses of 1369 (89.48%) beta thalassemia major cases were confirmed by the molecular testing. Some of the other commonly detected phenotypes were haemoglobin E-beta thalassemia in 43 individuals (2.81%), sickle cell disease in 40 individuals (2.61%) and sickle-beta thalassemia in 29 individuals (1.90%). Less common (< 1%) phenotypes detected included haemoglobin S-haemoglobin D (HbS/HbD) thalassemia (4 individuals), haemoglobin E-haemoglobin S (HbE/HbS) thalassemia (1 individual) and haemoglobin E homozygous (1 individual). Of the 43 cases with heterozygous HBB gene variations, the majority of individuals (thirty-six individuals) in our study were beta thalassemia carriers followed by five individuals harbouring the sickle cell trait, two individuals harbouring the HbE trait and one individual with a novel c.7delC heterozygous mutation (Table 1), which has been described subsequently.
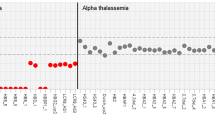
The NGS data included a spectrum of 48 pathogenic variants in the HBB gene (Fig. 1); of these, 45 variants were associated with beta thalassemia. The three other Hb variants detected were c.20A>T (HbS), c.79G>A (HbE) and c.364G>C (HbD Punjab) with allele frequencies of 4.18%, 1.66%, and 0.13%, respectively. The commonest pathogenic variants detected were the c.92+5G>C (IVS-I-5), the 619-bp deletion, c.92+1G>T, c.27_28insG, c.47G>A, c.126_129delCTTT, c.20A>T, and c.92G>C with allele frequencies of 44.55% (1344/3017 alleles), 10.74% (324/3017 alleles), 6.99% (211/3017 alleles), 6.23% (188/3017 alleles), 5.77% (174/3017 alleles), 4.71% (142/3017 alleles), 4.18% (126/3017 alleles) and 2.49% (75/3017 alleles), respectively (Fig. 1). Seven other mutations, c.51delC, c.79G>A, c.*110T>C, c.92+1G>A, c.17_18delCT, c.-50A>C and c.316-14T>G, had a combined allele frequency of 10.14% (306/3017 alleles). The remaining 33 pathogenic variants with individual frequencies of less than 1% accounted for 4.21% (127/3017 alleles) of the total allele frequencies (Supplementary Table S2). To ascertain whether the target assay can detect deep intronic variants in addition to the commonly encountered ones listed in this study, we mined our sequence data to search for cases that harboured previously described deep intronic variants in the intronic regions between exons 2 and 3. We have included a couple of examples in the file supplementary data 1.
Summary of the allelic frequencies of the beta thalassemia variants. *Others: Refer to Supplementary Table S2
For diagnostic yield calculations, clinically diagnosed cases of beta thalassemia and the other diagnosed haemoglobinopathies with biallelic pathogenic/likely pathogenic variants were considered to harbour two alleles whereas heterozygous cases/carriers were considered to harbour a single variant (Table 1).
Orthogonal testing by ARMS PCR, end-point PCR and capillary sequencing for the validation of our NGS assay
Our NGS assay was orthogonally tested with 136 samples; ARMS PCR was used to detect the c.92+5G>C mutation in 56 samples, end-point PCR was used to check for presence of the 619-bp deletion in 40 samples, and CE sequencing was used to verify the presence of the other mutations in 40 samples.
ARMS PCR: Concordance between ARMS PCR and NGS was 96.43% (54 out of 56 samples); the ARMS PCR and NGS data were discordant for a total of two samples (2/56). Two samples were negative for the c.92+5G>C mutation by ARMS PCR but were determined to be heterozygous for that mutation by NGS. A potential cause of error in the ARMS PCR results was the presence of other nucleotide variations in close proximity to the c.92 + 5 mutation within the PCR primer binding site, which were revealed by the NGS-based testing: a c.92+1G>T mutation in one sample and a c.92+1G>A mutation in the other (Fig. 2).
Comparison of ARMS PCR and NGS for detection of c.92+5G>C mutation. a Real-time ARMS PCR showing absence of the c.92+5G>C mutation. b Detection of the c.92+5G>C and c.92+1G>T heterozygous pathogenic variants by NGS. Visualization of the aligned BAM files in GenomeBrowse showing heterozygous pathogenic variants c.92+5G>C and c.92+1G>T that are located very close to each other. c Detection of the c.92+5G>C and c.92+1G>A heterozygous pathogenic variants by NGS. Visualization of the aligned BAM files in GenomeBrowse showing heterozygous pathogenic variants c.92+5G>C and c.92+1G>A that are located very close to each other
End-point PCR: The orthogonal testing for the 619-bp deletion showed 100% concordance between end-point PCR and NGS for 40 samples.
CE sequencing: Forty samples were orthogonally tested by CE sequencing for detection of mutations other than the c.92+5G>C mutation and the 619-bp deletion; the concordance between CE sequencing and NGS was 95% (38/40). CE sequencing was unable to unambiguously resolve the genotypes for two samples (the electropherograms exhibited “garbled” sequence data that could not be analysed), both of which were shown to harbour compound heterozygous mutations by NGS-based testing: a c.27_28insG heterozygous insertion and a c.51delC heterozygous deletion (Fig.3).
CE-seq (electropherograms) and NGS data of the c.27_28insG and c.51delC heterozygous deletions. a Visualization of the CE-seq forward sequence in 4peaks highlighting the beginning of the c.27_28insG mutation site. b Visualization of the CE-seq reverse complementary sequence in 4peaks highlighting the c.51delC mutation site. c Detection of the c.51delC and c.27_28insG heterozygous pathogenic variants by NGS. Visualization of the aligned BAM files in GenomeBrowse showing heterozygous pathogenic variants c.51delC and c.27_28insG
Novel mutation: We identified a novel variant, c.7delC, in three members of a single family. Two affected siblings harboured this mutation in a heterozygous state along with the c.92+5G>C mutation (Fig. 4), whereas the mother was an unaffected heterozygous carrier of the c.7delC mutation (with HPLC results indicating a beta thalassemia carrier status). This single-base deletion at nucleotide position 7 in the HBB gene is predicted to result in a frameshift termination following codon 3 (H3Ifs*2). This mutation has not been previously described in the literature. Hence, it was reported as a “variant of unknown significance (VUS), likely pathogenic.” It is likely to result in a B0 type of thalassemia as the frameshift will result in the production of an inactive polypeptide. We have submitted this novel variant to the HbVar database (HbVar ID 3193).
NGS images of the single-family harbouring the heterozygous c.7delC novel mutation. a Detection of the c.7delC and c.92+5G>C heterozygous pathogenic variants by NGS. Visualization of the aligned BAM files in GenomeBrowse of one of the affected siblings harbouring c.7delC novel mutation and c.92+5G>C in the compound heterozygous state. b Detection of the c.7delC and c.92+5G>C heterozygous pathogenic variants by NGS. Visualization of the aligned BAM files in GenomeBrowse of the other affected sibling harbouring a c.7delC novel mutation and c.92+5G>C in the compound heterozygous state. c Detection of the c.7delC heterozygous pathogenic variant by NGS. Visualization of the aligned BAM files in GenomeBrowse of the mother of the two affected siblings harbouring a c.7delC novel mutation in the heterozygous state. d Detection of the c.92+5G>C heterozygous pathogenic variant by NGS. Visualization of the aligned BAM files in GenomeBrowse of the father of the two affected siblings harbouring a c.92+5G>C heterozygous mutation
Discussion
We have developed a novel method to detect HBB gene mutations using a targeted NGS assay. This approach is an example of the potential of NGS as a cost-effective and practical tool for the analysis of single-gene disorders. The IVS-I-5 (c. 92+5G>C) mutation was the most commonly encountered pathogenic variant detected in our cohort. To validate the NGS results, we orthogonally tested samples from 136 patients using ARMS PCR, end-point PCR, and CE sequencing. The concordance between samples tested orthogonally by NGS and by other methodologies was 97.06%. In four cases, NGS was able to correctly identify the genotypes which were missed by ARMS PCR and CE sequencing.
Shah et al. analysed 75 referral samples of beta thalassemia from an east-western Indian population using ARMS PCRs (for the eight common Indian mutations: c.92+5G>C, the 619-bp deletion, c.79G>A, c.47G>A, c.364G>C, c.27_28insG, c.51delC and c.124_127delTTCT), capillary sequencing and end-point PCR. Their results showed that the most common mutation was the c.92+5 G>C (60.29%), followed by the 619-bp deletion (13.23%). Not surprisingly, these were the most abundant mutations in our cohort as well. However, for nearly two-thirds of their cohort [48 of 75 samples (64%)], multiple methods were needed to perform a complete analysis.
Many studies have shown that the mutation spectrum in the HBB gene varies geographically. For example, studies have shown that the IVS-I-6 [T>C] mutation is the most commonly encountered mutation in the Egyptian and Brazilian [10, 11] populations; the IVS-II-1 (G>A) mutation is the most common in the north of Iran and the IVS-I-5 (G>C) mutation in the south of Iran and Oman [12, 13]. As our assay does not target a limited subset of specific mutations, it is equally applicable to all geographies.
For ARMS PCR analysis, the mutation needs to be known a priori; however, this information is not required when using the targeted NGS assay to detect mutations. Additionally, ARMS PCR may yield false negative results, as shown by the two discrepant results observed in our cohort. Commonly encountered proximate mutations like the 92+5G>C and 92+1G>T mutations are likely to be missed by ARMS PCR if they are present in the same sample (Fig. 2). Another shortcoming of using PCR-based assays is allelic dropout in cases where polymorphisms at the 3′ end of the primer may cause only one allele of the gene to preferentially amplify [14]. Additionally, heterozygous deletions may be difficult to interpret using capillary sequencing as compared with interpretation by NGS. We have demonstrated that such mutation pairs can easily be identified by our targeted NGS assay. More than 80% of our HBB data had a Q-score ≥ 30 (which corresponds to less than one error per 1000 bases) with an average read depth greater than 300×. The higher number of reads is helpful in interpreting the data in an easy and unambiguous manner. Importantly, this NGS-based approach allows multiple classes of mutations to be analysed with equal ease. This is especially true for cases with multiple indels, which are amenable to simultaneous detection by NGS, as opposed to a sequential/multi-step analysis approach by CE-Seq.
An important utility of using a targeted NGS assay for thalassemia analysis is resolving ambiguous cases. All cases analysed in this study matched the genotype except for a heterozygous case with a thalassemia intermedia phenotype. This was the case of a 25-year-old female who presented with anaemia, moderate splenomegaly and mild haemolytic facies with no history of blood transfusions. The Hb HPLC results were suggestive of the individual being thalassemia minor. However, in view of the splenomegaly and peripheral blood smear findings suggesting haemolytic anaemia, HBB gene sequencing with MLPA for α thalassemia was performed. The patient harboured a c.17_18delCT heterozygous mutation in the HBB gene and a heterozygous triplication of the HBA gene (Supplementary figure S2). These data helped resolve the discrepancy between the clinical and HPLC findings.
Elucidation of mutations in the HBB gene is now becoming increasingly relevant from a therapeutic perspective as well. This is because many of these mutations can potentially be remedied in the not-too-distant future using a combination of gene editing technologies like the CRISPR/CAS9 system [15] and autologous stem cell transplantation. In fact, the CRISPR/CAS and older TALEN-based gene editing systems have already been used clinically to demonstrate the utility of gene editing for patients harbouring mutation such as the IVS-I-110(G>A) mutation [16].
A limitation of this study is the lack of complete demographic data for all samples tested in the present study. This data would have been useful to ascertain HBB variant trends and prevalence in specific areas and populations. Another perceived limitation is that this assay specifically targets the HBB gene only and may not detect large structural rearrangements that are not within the HBB gene. If there is a strong clinical suspicion of a large deletion (other than the 619-bp deletion) in the HBB gene cluster, then HBB MLPA analysis is recommended.
In summary, we have analysed the largest patient cohort of thalassemia and other haemoglobinopathies in India using a cost-effective indigenous NGS assay targeting the HBB gene. The assay is highly specific and sensitive. Within the context of our laboratory flow, which handles a large number of heterogeneous samples by NGS, the beta thalassemia assay is more economical and conducive to an integrated laboratory flow as compared with using a combination of ARMS PCR and CE sequencing. The cost of NGS library preparation is only marginally higher than that of the three sequential nested PCRs and reduces further with the level of multiplexing, which is limited only by the choice of sequencing platform and the number of barcodes present. For a different project, we have demonstrated multiplexing over 6000 samples per run of an Illumina NextSeq instrument. Similar or higher levels of multiplexing are possible with this assay, thereby making this approach significantly cheaper and more scalable than capillary sequencing–based assays. It is likely that other laboratories with a similar workflow will see an analogous benefit. Finally, the NGS assay is amenable to an extremely high degree of multiplexing, which can enable large-scale screening programs at costs comparable with HPLC/Hb electrophoresis.
References
Thacker N (2007) Prevention of thalassemia in India. Indian Pediatr 44:647–648
Cao A, Galanello R (2010) Beta-thalassemia. Genet Med 12:61–76
Varawalla NY, Old JM, Sarkar R, Venkatesan R, Weatherall DJ (1991) The spectrum of β-thalassaemia mutations on the Indian subcontinent: the basis for prenatal diagnosis [internet]. 78, Br J Haematol. 242–247. Available from: https://doi.org/10.1111/j.1365-2141.1991.tb04423.x.
Lin M, Zhu J-J, Wang Q, Xie L-X, Lu M, Wang J-L, Wang CF, Zhong TY, Zheng L, Pan MC, Wu JR, Wen YF, Liu GR, Zhan XF, Lin F, Yang LY (2012) Development and evaluation of a reverse dot blot assay for the simultaneous detection of common alpha and beta thalassemia in Chinese. Blood Cells Mol Dis 48:86–90
Fortina P, Dotti G, Conant R, Monokian G, Parrella T, Hitchcock W, Rappaport E, Schwartz E, Surrey S (1992) Detection of the most common mutations causing beta-thalassemia in Mediterraneans using a multiplex amplification refractory mutation system (MARMS). PCR Methods Appl 2:163–166
Shah PS, Shah ND, Ray HSP, Khatri NB, Vaghasia KK, Raval RJ, et al (2017) Mutation analysis of β-thalassemia in East-Western Indian population: a recent molecular approach [Internet]. Appl Clin Genet 10:27–35. Available from: https://doi.org/10.2147/tacg.s127531.
Wang W (2003) Multiplex Minisequencing screen for common Southeast Asian and Indian -thalassemia mutations [internet]. Clinical Chemistry 49:209–18. Available from: https://doi.org/10.1373/49.2.209,
Old JM, Varawalla NY, Weatherall DJ (1990) Rapid detection and prenatal diagnosis of beta-thalassaemia: studies in Indian and Cypriot populations in the UK. Lancet 336:834–837
Chan OTM, Westover KD, Dietz L, Zehnder JL, Schrijver I (2010) Comprehensive and efficient HBB mutation analysis for detection of beta-hemoglobinopathies in a pan-ethnic population. Am J Clin Pathol 133:700–707
Elmezayen AD, Kotb SM, Sadek NA, Abdalla EM (2015) β-Globin mutations in Egyptian patients with β-thalassemia. Lab Med 46:8–13
Silveira ZML da, da Silveira ZML, Barbosa M das V, de Medeiros Fernandes TAA, Kimura EM, Costa FF, et al (2011) Characterization of beta-thalassemia mutations in patients from the state of Rio Grande do Norte, Brazil [Internet]. Genet Mol Biol 34:425–8. Available from: https://doi.org/10.1590/s1415-47572011005000032.
Najmabadi H, Karimi-Nejad R, Sahebjam S, Pourfarzad F, Teimourian S, Sahebjam F, et al (2001) The β- thalassemia mutation spectrum in the iranian population [Internet]. Hemoglobin 25:285–96. Available from: https://doi.org/10.1081/hem-100105221.
Hassan SM, Hamza N, Al-Lawatiya FJ, Mohammed AJ, Harteveld CL, Rajab A, et al (2010) Extended molecular spectrum of β- and α-thalassemia in Oman [Internet], Hemoglobin. 34:127–34. Available from: https://doi.org/10.3109/03630261003673147.
Chehab FF (2010) Molecular diagnostic challenges of the thalassemias [Internet]. Molecular Diagnostics. p. 441–52. Available from: https://doi.org/10.1016/b978-0-12-369428-7.00036-7
Cai L, Bai H, Mahairaki V, Gao Y, He C, Wen Y, Jin YC, Wang Y, Pan RL, Qasba A, Ye Z, Cheng L (2018) A universal approach to correct various HBB gene mutations in human stem cells for gene therapy of beta-thalassemia and sickle cell disease. Stem Cells Transl Med 7:87–97
Patsali P, Turchiano G, Papasavva P, Romito M, Loucari CC, Stephanou C, Christou S, Sitarou M, Mussolino C, Cornu TI, Antoniou MN, Lederer CW, Cathomen T, Kleanthous M (2019) Correction of IVS I-110(G>A) β-thalassemia by CRISPR/Cas- and TALEN-mediated disruption of aberrant regulatory elements in human hematopoietic stem and progenitor cells [Internet]. Haematologica 104:e497–e501 Available from: https://doi.org/10.3324/haematol.2018.215178
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 12.7 kb)
ESM 2
(DOCX 14.6 kb)
ESM 3
(JPG 61.4 kb)
Supplementary Figure S1
Visualization of NGS data of samples harbouring the 619-bp deletion (NG_000007.3:g.71609_72227del619) in homozygous form, heterozygous form, and a sample without the deletion (PNG 1385 kb)
Supplementary Figure S2
NGS and MLPA findings of the patient harbouring a heterozygous c.17_18delCT mutation. (a) Detection of the c.17_18delCT heterozygous pathogenic variant by NGS. Visualization of the aligned BAM files in GenomeBrowse showing heterozygous pathogenic variant c.17_18delCT. (b) MLPA result of the patient indicating a triplication in the HBA gene (PNG 4077 kb)
ESM 4
(DOC 279 kb)
Rights and permissions
About this article
Cite this article
Kelkar, K., Ramanan, V., Anand, S. et al. HBB gene mutation spectrum in an Indian cohort of 1530 cases using an in-house targeted next-generation sequencing assay. J Hematopathol 13, 239–248 (2020). https://doi.org/10.1007/s12308-020-00414-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12308-020-00414-8