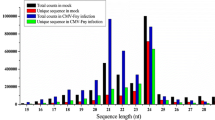
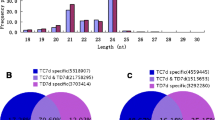
Abstract
Increasing incidence of viral infections in crop plants adversely affects their growth and yield. Tomato (Solanum lycopersicum) is considered to be a favorite host for viruses with over 50 species of begomoviruses naturally infecting this crop. Tomato leaf curl virus (ToLCV) is among the most widespread and devastating begomoviruses affecting tomato production. microRNAs (miRs) have been established as key regulators of gene expression and plant development. The miR pathways are disturbed during infection by viruses. Thus, comprehension of regulatory miR networks is crucial in understanding the effect of viral pathogenicity. To identify key miRs involved in ToLCV infection, a high throughput approach involving next generation sequencing was employed. Healthy and infected leaf tissues of two tomato varieties, differing in their susceptibility to ToLCV infection were analyzed. NGS data analysis followed by computational predictions, led to identification of 91 known miRs, 15 novel homologs and 53 novel miRs covering two different varieties of tomato, susceptible (Pusa Ruby) and tolerant (LA1777) to ToLCV infection. The cleaved targets of these miRs were identified using online available degradome libraries from leaf, flower and fruit of tomato and showed their involvement in various biological pathways through KEGG Orthology. With detailed comparative profiling of expression pattern of these miRs, we could associate the specific miRs with the resistant and infected genotypes. This study depicted that in depth analysis of miR expression patterns and their functions will help in identification of molecules that can be used for manipulation of gene expression to increase crop production and developing resistance against diseases.







Similar content being viewed by others
References
Achard P, Herr A, Baulcombe DC, Harberd NP (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131(14):3357–3365. https://doi.org/10.1242/dev.01206
Addo-Quaye C, Miller W, Axtell MJ (2009) CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25(1):130–131. https://doi.org/10.1093/bioinformatics/btn604
Alazem M, Lin NS (2015) Roles of plant hormones in the regulation of host–virus interactions. Mol Plant Pathol 16(5):529–540. https://doi.org/10.1111/mpp.12204
Azzam O, Frazer J, De La Rosa D, Beaver JS, Ahlquist P, Maxwell DP (1994) Whitefly transmission and efficient ssDNA accumulation of bean golden mosaic geminivirus require functional coat protein. Virology 204(1):289–296. https://doi.org/10.1006/viro.1994.1533
Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA (2009) NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res 37(suppl 1):D885–D890. https://doi.org/10.1093/nar/gkn764
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. https://doi.org/10.1016/S0092-8674(04)00045-5
Bonnet E, Wuyts J, Rouzé P, Van de Peer Y (2004) Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20(17):2911–2917. https://doi.org/10.1093/bioinformatics/bth374
Boulton MI (2003) Geminiviruses: major threats to world agriculture. Ann Appl Biol 142(2):143–143. https://doi.org/10.1111/j.1744-7348.2003.tb00239.x
Briddon RW, Mansoor S, Bedford ID, Pinner MS, Markham PG (2000) Clones of cotton leaf curl geminivirus induce symptoms atypical of cotton leaf curl disease. Virus Genes 20(1):19–26. https://doi.org/10.1023/A:1008151921937
Carbonell A, Carrington JC (2015) Antiviral roles of plant ARGONAUTES. Curr Opin Plant Biol 27:111–117. https://doi.org/10.1016/j.pbi.2015.06.013
Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303(5666):2022–2025. https://doi.org/10.1126/science.1088060
Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang H-S, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14(3):559–574. https://doi.org/10.1105/tpc.010410
Chen T, Lv Y, Zhao T, Li N, Yang Y, Yu W, He X, Liu T, Zhang B (2013) Comparative transcriptome profiling of a resistant vs. susceptible tomato (Solanum lycopersicum) cultivar in response to infection by tomato yellow leaf curl virus. PLoS ONE 8(11):e80816. https://doi.org/10.1371/journal.pone.0080816
Chinnusamy V, Zhu J-K (2009) RNA-directed DNA methylation and demethylation in plants. Sci China, Ser C Life Sci 52(4):331–343. https://doi.org/10.1007/s11427-009-0052-1
Cui X, Xu SM, Mu DS, Yang ZM (2009) Genomic analysis of rice microRNA promoters and clusters. Gene 431(1–2):61–66. https://doi.org/10.1016/j.gene.2008.11.016
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2(2):e219. https://doi.org/10.1371/journal.pone.0000219
Guo H-S, Xie Q, Fei J-F, Chua N-H (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell Online 17(5):1376–1386. https://doi.org/10.1105/tpc.105.030841
Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S (2013) Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol 11(11):777–788. https://doi.org/10.1038/nrmicro3117
Huang Y, Zhang B-L, Sun S, Xing G-M, Wang F, Li M-Y, Tian Y-S, Xiong A-S (2016) AP2/ERF transcription factors involved in response to Tomato yellow leaf curly virus in Tomato. Plant Genome. https://doi.org/10.3835/plantgenome2015.09.0082
Hunter W, Hiebert E, Webb S, Tsai J, Polston J (1998) Location of geminiviruses in the whitefly Bemisia tabaci (Homoptera: Aleyrodidae). Plant Dis 82(10):1147–1151. https://doi.org/10.1094/PDIS.1998.82.10.1147
Huntley RP, Sawford T, Mutowo-Meullenet P, Shypitsyna A, Bonilla C, Martin MJ, O’Donovan C (2015) The GOA database: gene Ontology annotation updates for 2015. Nucleic Acids Res 43:d1057–1063. https://doi.org/10.1093/nar/gku1113
Ingham DJ, Pascal E, Lazarowitz SG (1995) Both bipartite geminivirus movement proteins define viral host range, but only BL1 determines viral pathogenicity. Virology 207(1):191–204. https://doi.org/10.1006/viro.1995.1066
Jiao Y, Song W, Zhang M, Lai J (2011) Identification of novel maize miRNAs by measuring the precision of precursor processing. BMC Plant Biol 11:141. https://doi.org/10.1186/1471-2229-11-141
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14(6):787–799. https://doi.org/10.1016/j.molcel.2004.05.027
Kidner CA, Martienssen RA (2004) Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428(6978):81–84. https://doi.org/10.1038/nature02366
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research 42:D68–D73. https://doi.org/10.1093/nar/gkt1181 (Database issue)
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3):R25. https://doi.org/10.1186/gb-2009-10-3-r25
Li C-W, Su R-C, Cheng C-P, You S-J, Hsieh T-H, Chao T-C, Chan M-T (2011) Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol 156(1):213–227. https://doi.org/10.1104/pp.111.174268
Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B (2012) MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 109(5):1790–1795. https://doi.org/10.1073/pnas.1118282109
Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199(3):639–649. https://doi.org/10.1111/nph.12291
Lorenz R, Bernhart SH, Zu Siederdissen CH, Tafer h, Flamm C, Stadler PF, Hofacker IL (2011) ViennaRNA package 2.0. Algorithms. Mol Biol 6:26. https://doi.org/10.1186/1748-7188-6-26
Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 23(16):3356–3364. https://doi.org/10.1038/sj.emboj.7600340
Mansoor S, Briddon RW, Zafar Y, Stanley J (2003) Geminivirus disease complexes: an emerging threat. Trends in Plant Science 8(3):128–134. https://doi.org/10.1016/S1360-1385(03)00007-4
Martinez G, Forment J, Llave C, Pallas V, Gomez G (2011) High-throughput sequencing, characterization and detection of new and conserved cucumber miRNAs. PLoS ONE 6(5):e19523. https://doi.org/10.1371/journal.pone.0019523
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martienssen RA, Poethig RS, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhu JK (2008) Criteria for annotation of plant MicroRNAs. Plant Cell 20(12):3186–3190. https://doi.org/10.1105/tpc.108.064311
Moffat AS (1999) Geminiviruses emerge as serious crop threat. Science 286(5446):1835–1835
Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ (2008) Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res 18(4):571–584. https://doi.org/10.1101/gr.6897308
Moriones E, Navas-Castillo J (2000) Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res 71(1–2):123–134. https://doi.org/10.1016/S0168-1702(00)00193-3
Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res 18(10):1602–1609. https://doi.org/10.1101/gr.080127.108
Naqvi AR, Islam MN, Choudhury NR, Haq QM (2009) The fascinating world of RNA interference. Int J Biol Sci 5(2):97–117. https://doi.org/10.7150/ijbs.5.97
Naqvi AR, Haq Q, Mukherjee SK (2010) MicroRNA profiling of Tomato leaf curl New Delhi virus (ToLCNDV) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol J 7(1):281. https://doi.org/10.1186/1743-422X-7-281
Naqvi AR, Choudhury NR, Mukherjee SK, Haq QMR (2011) In silico analysis reveals that several tomato microRNA/microRNA* sequences exhibit propensity to bind to tomato leaf curl virus (ToLCV) associated genomes and most of their encoded open reading frames (ORFs). Plant Physiol Biochem 49(1):13–17. https://doi.org/10.1016/j.plaphy.2010.09.013
Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell Online 18(11):2929–2945. https://doi.org/10.1105/tpc.106.045617
Nosaka M, Ishiwata A, Shimizu-Sato S, Ono A, Ishimoto K, Noda Y, Sato Y (2013) The copy number of rice CACTA DNA transposons carrying MIR820 does not correlate with MIR820 expression. Plant Signal Behav 8(8):e25169. https://doi.org/10.4161/psb.25169
Nuruzzaman M, Sharoni AM, Kikuchi S (2013) Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol 4:248. https://doi.org/10.3389/fmicb.2013.00248
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425(6955):257–263. https://doi.org/10.1038/nature01958
Park J, Lee H-J, Cheon C-I, Kim S-H, Hur Y-S, Auh C-K, Im K-H, Yun D-J, Lee S, Davis KR (2011) The Arabidopsis thaliana homeobox gene ATHB12 is involved in symptom development caused by geminivirus infection. PLoS ONE 6(5):e20054. https://doi.org/10.1371/journal.pone.0020054
Pascal E, Sanderfoot AA, Ward BM, Medville R, Turgeon R, Lazarowitz SG (1994) The geminivirus BR1 movement protein binds single-stranded DNA and localizes to the cell nucleus. Plant Cell 6(7):995–1006. https://doi.org/10.1105/tpc.6.7.995
Peng T, Lv Q, Zhang J, Li J, Du Y, Zhao Q (2011) Differential expression of the microRNAs in superior and inferior spikelets in rice (Oryza sativa). J Exp Bot 62(14):4943–4954. https://doi.org/10.1093/jxb/err205
Pradhan B, Naqvi AR, Saraf S, Mukherjee SK, Dey N (2015) Prediction and characterization of Tomato leaf curl New Delhi virus (ToLCNDV) responsive novel microRNAs in Solanum lycopersicum. Virus Res 195:183–195. https://doi.org/10.1016/j.virusres.2014.09.001
Puzey JR, Karger A, Axtell M, Kramer EM (2012) Deep annotation of Populus trichocarpa microRNAs from diverse tissue sets. PLoS ONE 7(3):e33034. https://doi.org/10.1371/journal.pone.0033034
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110(4):513–520. https://doi.org/10.1016/S0092-8674(02)00863-2
Rigden JE, Krake LR, Rezaian MA, Dry IB (1994) ORF C4 of tomato leaf curl geminivirus is a determinant of symptom severity. Virology 204(2):847–850. https://doi.org/10.1006/viro.1994.1606
Saraf S, Sanan-Mishra N, Gursanscky NR, Carroll BJ, Gupta D, Mukherjee SK (2015) 3′ and 5′ microRNA-end post-biogenesis modifications in plant transcriptomes: evidences from small RNA next generation sequencing data analysis. Biochem Biophys Res Commun 467(4):892–899. https://doi.org/10.1016/j.bbrc.2015.10.048
Seal S, VandenBosch F, Jeger M (2006) Factors influencing begomovirus evolution and their increasing global significance: implications for sustainable control. Crit Rev Plant Sci 25(1):23–46. https://doi.org/10.1080/07352680500365257
Sharma N, Tripathi A, Sanan-Mishra N (2015) Profiling the expression domains of a rice-specific microRNA under stress. Front Plant Sci 6:333. https://doi.org/10.3389/fpls.2015.00333
Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, Moulton V, Dalmay T (2008) High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genom 9:593. https://doi.org/10.1186/1471-2164-9-593
Tripathi A, Goswami K, Sanan-Mishra N (2015) Role of bioinformatics in establishing microRNAs as modulators of abiotic stress responses: the new revolution. Front Physiol. https://doi.org/10.3389/fphys.2015.00286
Voorrips R (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78. https://doi.org/10.1093/jhered/93.1.77
Wang W, Luan Y (2015) The advance of tomato disease-related microRNAs. Plant Cell Rep 34(7):1089–1097. https://doi.org/10.1007/s00299-015-1782-0
Wang Y, Hu Z, Yang Y, Chen X, Chen G (2009) Function annotation of an SBP-box gene in Arabidopsis based on analysis of co-expression networks and promoters. Int J Mol Sci 10(1):116–132. https://doi.org/10.3390/ijms10010116
Wang J, Yu W, Yang Y, Li X, Chen T, Liu T, Ma N, Yang X, Liu R, Zhang B (2015) Genomewide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci Rep 18(5):16946. https://doi.org/10.1038/srep16946
Warner JR, McIntosh KB (2009) How common are extraribosomal functions of ribosomal proteins? Mol Cell 34(1):3–11. https://doi.org/10.1016/j.molcel.2009.03.006
Yang X, Li L (2011) miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics 27(18):2614–2615. https://doi.org/10.1093/bioinformatics/btr430
Yang C, Zhang C, Dittman JD, Whitham SA (2009) Differential requirement of ribosomal protein S6 by plant RNA viruses with different translation initiation strategies. Virology 390(2):163–173. https://doi.org/10.1016/j.virol.2009.05.018
Yang X, Guo W, Ma X, An Q, Zhou X (2011) Molecular characterization of Tomato leaf curl China virus, infecting tomato plants in China, and functional analyses of its associated betasatellite. Appl Environ Microbiol 77(9):3092–3101. https://doi.org/10.1128/AEM.00017-11
Zhai J, Jeong D-H, De Paoli E, Park S, Rosen BD, Li Y, González AJ, Yan Z, Kitto SL, Grusak MA (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25(23):2540–2553. https://doi.org/10.1101/gad.177527.111
Zhang W, Gao S, Zhou X, Xia J, Chellappan P, Zhou X, Zhang X, Jin H (2010) Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol 11(8):R81. https://doi.org/10.1186/gb-2010-11-8-r81
Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, Luo J (2011) PlantTFDB 2.0: Update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res 39(suppl 1):D1114–D1117. https://doi.org/10.1093/nar/gkq1141
Zhao W, Li Z, Fan J, Hu C, Yang R, Qi X, Chen H, Zhao F, Wang S (2015) Identification of jasmonic acid-associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root-knot nematode stress in tomato. J Exp Bot 66(15):4653–4667. https://doi.org/10.1093/jxb/erv238
Acknowledgements
This research was supported by grants from Department of Biotechnology, New Delhi, India. The authors acknowledge the efforts of Dr. Afsar Raza Naqvi in preparing the tomato sRNA libraries.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
12298_2017_482_MOESM1_ESM.xlsx
Supplementary File 1: Lists all the predicted novel miRs, their precursors, genomic loci, associated family and isoforms
12298_2017_482_MOESM2_ESM.pdf
Supplementary File 2: Secondary structure of all predicted miRs and their isoforms. miR is highlighted with blue and miR* is highlighted with red color and the circle at the last base represents 5′ end of the precursor
12298_2017_482_MOESM4_ESM.jpg
Supplementary File 4: Validation of miRs by stem-loop RT-PCR in tissues infiltrated with agrobacterium clone. Lane 1: Low range DNA ladder (25 bp), 7 s: shoot tissue 7 days post infiltration, 7 r: root tissue 7 days post infiltration, 28 s: shoot tissue 28 days post infiltration, 28r: root tissue 28 days post infiltration
12298_2017_482_MOESM6_ESM.pdf
Supplementary File 6: Target-plots (t-plots) and cleavage sites of all the degradome validated targets. Red dot indicates signatures consistent with miR-directed cleavage site. The abundance of each signature is plotted as a function of its position in the transcript
Rights and permissions
About this article
Cite this article
Tripathi, A., Goswami, K., Tiwari, M. et al. Identification and comparative analysis of microRNAs from tomato varieties showing contrasting response to ToLCV infections. Physiol Mol Biol Plants 24, 185–202 (2018). https://doi.org/10.1007/s12298-017-0482-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-017-0482-3