Abstract
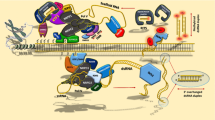
RNA-directed DNA methylation (RdDM) is a nuclear process in which small interfering RNAs (siRNAs) direct the cytosine methylation of DNA sequences that are complementary to the siRNAs. In plants, double stranded-RNAs (dsRNAs) generated by RNA-dependent RNA polymerase 2 (RDR2) serve as precursors for Dicer-like 3 dependent biogenesis of 24-nt siRNAs. Plant specific RNA polymerase IV (Pol IV) is presumed to generate the initial RNA transcripts that are substrates for RDR2. siRNAs are loaded onto an argonaute4-containing RISC (RNA-induced silencing complex) that targets the de novo DNA methyltransferase DRM2 to RdDM target loci. Nascent RNA transcripts from the target loci are generated by another plant-specific RNA polymerase, Pol V, and these transcripts help recruit complementary siRNAs and the associated RdDM effector complex to the target loci in a transcription-coupled DNA methylation process. Small RNA binding proteins such as ROS3 may direct target-specific DNA demethylation by the ROS1 family of DNA demethylases. Chromatin remodeling enzymes and histone modifying enzymes also participate in DNA methylation and possibly demethylation. One of the well studied functions of RdDM is transposon silencing and genome stability. In addition, RdDM is important for paramutation, imprinting, gene regulation, and plant development. Locus-specific DNA methylation and demethylation, and transposon activation under abiotic stresses suggest that RdDM is also important in stress responses of plants. Further studies will help illuminate the functions of RdDM in the dynamic control of epigenomes during development and environmental stress responses.
Similar content being viewed by others
References
Cokus S J, Feng S, Zhang X, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature, 2008, 452: 215–219 18278030, 10.1038/nature06745, 1:CAS:528:DC%2BD1cXjt1GnurY%3D
Huettel B, Kanno T, Daxinger L, et al. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta, 2007, 1769: 358–374 17449119, 1:CAS:528:DC%2BD2sXmtlSnsb8%3D
Matzke M, Kanno T, Huettel B, et al. Targets of RNA-directed DNA methylation, Curr Opin Plant Biol, 2007, 10: 512–519 17702644, 10.1016/j.pbi.2007.06.007, 1:CAS:528:DC%2BD2sXhtFekurjL
Zhang X. The epigenetic landscape of plants. Science, 2008, 320: 489–492 18436779, 10.1126/science.1153996, 1:CAS:528:DC%2BD1cXkvFGktrw%3D
Zilberman D. The evolving functions of DNA methylation. Curr Opin Plant Biol, 2008, 11: 554–559 18774331, 10.1016/j.pbi.2008.07.004, 1:CAS:528:DC%2BD1cXhtFCnsr7E
Zhu J K. Epigenome sequencing comes of age. Cell, 2008, 133: 395–397 18455978, 10.1016/j.cell.2008.04.016, 1:CAS:528:DC%2BD1cXmt1SmtLk%3D
Lister R, O’Malley R C, Tonti-Filippini J, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell, 2008, 133: 523–536 18423832, 10.1016/j.cell.2008.03.029, 1:CAS:528:DC%2BD1cXmt1Smur8%3D
Wassenegger M, Heimes S, Riedel L, et al. RNA-directed de novo methylation of genomic sequences in plants. Cell, 1994, 76: 567–576 8313476, 10.1016/0092-8674(94)90119-8, 1:CAS:528:DyaK2cXis1artrk%3D
Park Y D, Papp I, Moscone E A, et al. Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J, 1996, 9: 183–194 8820605, 10.1046/j.1365-313X.1996.09020183.x, 1:CAS:528:DyaK28XitVyjur0%3D
Mette M F, Aufsatz W, van der Winden J, et al. Transcriptional gene silencing and promoter methylation triggered by double stranded RNA. EMBO J, 2000, 19: 5194–5201 11013221, 10.1093/emboj/19.19.5194, 1:CAS:528:DC%2BD3cXnvF2qtLk%3D
Hamilton A J, Baulcombe D C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science, 1999, 286: 950–952 10542148, 10.1126/science.286.5441.950, 1:CAS:528:DyaK1MXntFaktbY%3D
Jones-Rhoades M W, Bartel D P, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol, 2006, 57: 19–53 16669754, 10.1146/annurev.arplant.57.032905.105218, 1:CAS:528:DC%2BD28XosVKhsb0%3D
Mallory A C, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat Genet, 2006, 38: S31–S36 16736022, 10.1038/ng1791, 1:CAS:528:DC%2BD28XltVOmt7k%3D
Henderson I R, Jacobsen S E. Epigenetic inheritance in plants. Nature, 2007, 447: 418–424 17522675, 10.1038/nature05917, 1:CAS:528:DC%2BD2sXlsFOgs78%3D
Xie Z, Johansen L K, Gustafson A M, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol, 2004, 2(5):E104. doi:10.1371/journal.pbio.0020104
He X J, Hsu Y F, Pontes O, et al. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerase IV and V and is required for RNA-dicrected DNA methylation. Genes Dev, 2009, 23: 318–330 19204117, 10.1101/gad.1765209, 1:CAS:528:DC%2BD1MXit1KgtLw%3D
Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev, 2006, 20: 759–771 16600909, 10.1101/gad.1410506, 1:CAS:528:DC%2BD28XjsFGjsr0%3D
Pikaard C S, Haag J R, Ream T, et al. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci, 2008, 13: 390–397 18514566, 10.1016/j.tplants.2008.04.008, 1:CAS:528:DC%2BD1cXotlOjtL8%3D
Ream T S, Haag J R, Wierzbicki A T, et al. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell, 2008, 33: 192–203 19110459, 10.1016/j.molcel.2008.12.015, 1:CAS:528:DC%2BD1MXhvVylsbk%3D
Hamilton A, Voinnet O, Chappell L, et al. Two classes of short interfering RNA in RNA silencing. EMBO J, 2002, 21: 4671–4679 12198169, 10.1093/emboj/cdf464, 1:CAS:528:DC%2BD38Xms1KmsrY%3D
Kanno T, Mette M F, Kreil D P, et al. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol, 2004, 14: 801–805 15120073, 10.1016/j.cub.2004.04.037, 1:CAS:528:DC%2BD2cXjs1ejur0%3D
Kanno T, Huettel B, Mette M F, et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet, 2005, 37: 761–765 15924141, 10.1038/ng1580, 1:CAS:528:DC%2BD2MXlslWhsrc%3D
Herr A J, Jensen M B, Dalmay T, et al. RNA polymerase IV directs silencing of endogenous DNA. Science, 2005, 308: 118–120 15692015, 10.1126/science.1106910, 1:CAS:528:DC%2BD2MXivVaqsrw%3D
Onodera Y, Onodera Y, Haag J R, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell, 2005, 120: 613–622 15766525, 10.1016/j.cell.2005.02.007, 1:CAS:528:DC%2BD2MXis1yhsb0%3D
Pontier D, Yahubyan G, Vega D, et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev, 2005, 19: 2030–2040 16140984, 10.1101/gad.348405, 1:CAS:528:DC%2BD2MXhtVSlurnO
Li C F, Pontes O, El-Shami M, et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell, 2006, 126: 93–106 16839879, 10.1016/j.cell.2006.05.032, 1:CAS:528:DC%2BD28Xns1Cgt74%3D
Pontes O, Li C F, Nunes P C, et al. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell, 2006, 126: 79–92 16839878, 10.1016/j.cell.2006.05.031, 1:CAS:528:DC%2BD28Xns1Cgtrc%3D
Lahmy S, Pontier D, Cavel E, et al. PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc Natl Acad Sci USA, 2009, 106: 941–946 19141635, 10.1073/pnas.0810310106, 1:CAS:528:DC%2BD1MXksFGhtg%3D%3D
Huang L, Jones A M, Searle I, et al. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol, 2009, 16: 91–93 19079263, 10.1038/nsmb.1539, 1:CAS:528:DC%2BD1cXhsV2hsrbI
Zhang X, Henderson I R, Lu C, et al. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci USA, 2007, 104: 4536–4541 17360559, 10.1073/pnas.0611456104, 1:CAS:528:DC%2BD2sXjsFKqtbY%3D
Smith L M, Pontes O, Searle I, et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell, 2007, 19: 1507–1521 17526749, 10.1105/tpc.107.051540, 1:CAS:528:DC%2BD2sXnvVWqtL8%3D
Haag J R, Pontes O, Pikaard C S. Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing. PLoS ONE, 2009, 4(1): e4110
Cao X, Jacobsen S E. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA, 2002, 99(Suppl 4): 16491–16498 12151602, 10.1073/pnas.162371599, 1:CAS:528:DC%2BD38Xps12ht7c%3D
Zaratiegui M, Irvine D V, Martienssen R A. Noncoding RNAs and gene silencing. Cell, 2007, 128: 763–776 17320512, 10.1016/j.cell.2007.02.016, 1:CAS:528:DC%2BD2sXis12isr0%3D
Jones L, Ratcliff F, Baucombe D C. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol, 2001, 11: 747–757 11378384, 10.1016/S0960-9822(01)00226-3, 1:CAS:528:DC%2BD3MXjvFSiuro%3D
Cao X, Aufsatz W, Zilberman D, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol, 2003, 13: 2212–2217 14680640, 10.1016/j.cub.2003.11.052, 1:CAS:528:DC%2BD3sXhtVSjsrnM
Chan S W, Zilberman D, Xie Z, et al. RNA silencing genes control de novo DNA methylation. Science, 2004, 303: 1336 14988555, 10.1126/science.1095989, 1:CAS:528:DC%2BD2cXhslCru70%3D
Vaucheret H. Plant ARGONAUTES. Trends Plant Sci, 2008, 13: 350–358 18508405, 10.1016/j.tplants.2008.04.007, 1:CAS:528:DC%2BD1cXotlOjt7g%3D
Zilberman D, Cao X, Jacobsen S E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science, 2003, 299: 716–719 12522258, 10.1126/science.1079695, 1:CAS:528:DC%2BD3sXntFWhsg%3D%3D
Chan S W, Henderson I R, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet, 2005, 6: 351–360 15861207, 10.1038/nrg1601, 1:CAS:528:DC%2BD2MXjs1OksL0%3D
Matzke M A, Birchler J A. RNAi-mediated pathways in the nucleus. Nat Rev Genet, 2005, 6: 24–35 15630419, 10.1038/nrg1500, 1:CAS:528:DC%2BD2MXlvVGh
Qi Y, He X, Wang X J, et al. Distinct catalytic and non-catalytic roles of Argonaute4 in RNA-directed DNA methylation. Nature, 2006, 443: 1008–1012 16998468, 10.1038/nature05198
Zheng X, Zhu J, Kapoor A, et al. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J, 2007, 26: 1691–1701 17332757, 10.1038/sj.emboj.7601603, 1:CAS:528:DC%2BD2sXjt1WksbY%3D
Aufsatz W, Mette M F, van der Winden J, et al. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J, 2002, 21: 6832–6841 12486004, 10.1093/emboj/cdf663, 1:CAS:528:DC%2BD38XpslOhu74%3D
Ebbs M L, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell, 2006, 18: 1166–1176 16582009, 10.1105/tpc.106.041400, 1:CAS:528:DC%2BD28XkslCmurc%3D
Lindroth A M, Shultis D, Jasencakova Z, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J, 2004, 23: 4286–4296 15457214, 10.1038/sj.emboj.7600430, 1:CAS:528:DC%2BD2cXovFalsLw%3D
Ebbs M L, Bartee L, Bender J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Cell Biol, 2005, 25: 10507–10515 16287862, 10.1128/MCB.25.23.10507-10515.2005, 1:CAS:528:DC%2BD2MXht1OnsL%2FF
Johnson L M, Law J A, Khattar A, et al. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet, 2008, 4(11): e1000280
Sridhar V V, Kapoor A, Zhang K, et al. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature, 2007, 447: 735–738 17554311, 10.1038/nature05864, 1:CAS:528:DC%2BD2sXmtFels7w%3D
Jeddeloh J A, Stokes T L, Richards E J. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet, 1999, 22: 94–97 10319870, 10.1038/8803, 1:CAS:528:DyaK1MXjtVagsLo%3D
Lippman Z, Gendrel A V, Black M, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature, 2004, 430: 471–476 15269773, 10.1038/nature02651, 1:CAS:528:DC%2BD2cXlvVaksLk%3D
Chen H, Samadder P P, Tanaka Y, et al. OsRecQ1, a QDE-3 homologue in rice, is required for RNA silencing induced by particle bombardment with inverted repeat DNA, but not with double-stranded RNA. Plant J, 2008, 56: 274–286 18564381, 10.1111/j.1365-313X.2008.03587.x, 1:CAS:528:DC%2BD1cXhtlChu7rJ
Kanno T, Bucher E, Daxinger L, et al. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet, 2008, 40: 670–675 18425128, 10.1038/ng.119, 1:CAS:528:DC%2BD1cXltFylurk%3D
Heard E, Colot V. Chromosome structural proteins and RNA-mediated epigenetic silencing. Dev Cell, 2008, 14: 813–814 18539109, 10.1016/j.devcel.2008.05.013, 1:CAS:528:DC%2BD1cXnsVCrs78%3D
Mosher R A, Schwach F, Studholme D, et al. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci USA, 2008, 105: 3145–3150 18287047, 10.1073/pnas.0709632105
El-Shami M, Pontier D, Lahmy S, et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev, 2007, 21: 2539–2544 17938239, 10.1101/gad.451207, 1:CAS:528:DC%2BD2sXht1WnsbjN
Li C F, Henderson I R, Song L, et al. Dynamic regulation of ARGONAUTE4 within multiple nuclear bodies in Arabidopsis thaliana. PLoS Genet, 2008, 4(2): e27. doi:10.1371/journal.pgen. 0040027
Wierzbicki A T, Haag J R, Pikaard C S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell, 2008, 135: 635–648 19013275, 10.1016/j.cell.2008.09.035, 1:CAS:528:DC%2BD1cXhsVCls7rE
Daxinger L, Kanno T, Bucher E, et al. A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. EMBO J, 2009, 28: 48–57 19078964, 10.1038/emboj.2008.260, 1:CAS:528:DC%2BD1cXhsVylsb7E
Gong Z, Morales-Ruiz T, Ariza R R, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell, 2002, 111: 803–814 12526807, 10.1016/S0092-8674(02)01133-9, 1:CAS:528:DC%2BD3sXhtVyltA%3D%3D
Agius F, Kapoor A, Zhu J K. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA, 2006, 103: 11796–11801 16864782, 10.1073/pnas.0603563103, 1:CAS:528:DC%2BD28XotFansrk%3D
Choi Y, Gehring M, Johnson L, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell, 2002, 110: 33–42 12150995, 10.1016/S0092-8674(02)00807-3, 1:CAS:528:DC%2BD38XlsV2mt78%3D
Penterman J, Zilberman D, Huh J H, et al. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA, 2007, 104: 6752–6757 17409185, 10.1073/pnas.0701861104, 1:CAS:528:DC%2BD2sXkvFWjsbg%3D
Zhu J, Kapoor A, Sridhar V V, et al. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol, 2007, 17: 54–59 17208187, 10.1016/j.cub.2006.10.059, 1:CAS:528:DC%2BD2sXis1antg%3D%3D
Douet J, Blanchard B, Cuvillier C, et al Interplay of RNA Pol IV and ROS1 during post-embryonic 5S rDNA chromatin remodeling. Plant Cell Physiol, 2008, 49: 1783–1791 18845569, 10.1093/pcp/pcn152, 1:CAS:528:DC%2BD1MXptFSlsw%3D%3D
Zheng X, Pontes O, Zhu J, et al. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature, 2008, 455: 1259–1262 18815596, 10.1038/nature07305, 1:CAS:528:DC%2BD1cXhtlSku7fJ
Zhang X, Yazaki J, Sundaresan A, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell, 2006, 126: 1189–1201 16949657, 10.1016/j.cell.2006.08.003, 1:CAS:528:DC%2BD28XhtVCnsrvP
Zilberman D, Gehring M, Tran R K, et al. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet, 2007, 39: 61–69 17128275, 10.1038/ng1929, 1:CAS:528:DC%2BD28XhtlGktLvJ
Hirochika H, Okamoto H, Kakutani T. Silencing of retrotransposons in Arabidopsis and reactivation by the ddm1 mutation. Plant Cell, 2000, 12: 357–369 10715322, 10.1105/tpc.12.3.357, 1:CAS:528:DC%2BD3cXktFWjurc%3D
Miura A, Yonebayashi S, Watanabe K, et al. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature, 2001, 411: 212–214 11346800, 10.1038/35075612, 1:CAS:528:DC%2BD3MXjslGlurg%3D
Preuss S B, Costa-Nunes P, Tucker S, et al. Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol Cell, 2008, 32: 673–684 19061642, 10.1016/j.molcel.2008.11.009, 1:CAS:528:DC%2BD1cXhsFSntrrP
Zhai J, Liu J, Liu B, et al. Small RNA-directed epigenetic natural variation in Arabidopsis thaliana. PLoS Genet, 2008, 4(4): e1000056. doi:10.1371/ journal. pgen.1000056
Huettel B, Kanno T, Daxinger L, et al. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J, 2006, 25: 2828–2836 16724114, 10.1038/sj.emboj.7601150, 1:CAS:528:DC%2BD28XlvFKhs74%3D
Shibuya K, Fukushima S, Takatsuji H. RNA-directed DNA methylation induces transcriptional activation in plants. Proc Natl Acad Sci USA, 2009, 106: 1660–1665 19164525, 10.1073/pnas.0809294106
Alleman M, Sidorenko L, McGinnis K, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature, 2006, 442: 295–448 16855589, 10.1038/nature04884, 1:CAS:528:DC%2BD28XmvF2jsrg%3D
Chandler V L. Paramutation: from maize to mice. Cell, 2007, 128: 641–645 17320501, 10.1016/j.cell.2007.02.007, 1:CAS:528:DC%2BD2sXis12ju7w%3D
Erhard K F Jr, Stonaker J L, Parkinson S E, et al. RNA polymerase IV functions in paramutation in Zea mays. Science, 2009, 323: 1201–1205 19251626, 10.1126/science.1164508, 1:CAS:528:DC%2BD1MXit1eltLc%3D
Kurihara Y, Matsui A, Kawashima M, et al. Identification of the candidate genes regulated by RNA-directed DNA methylation in Arabidopsis. Biochem Biophys Res Commun, 2008, 376: 553–557 18805399, 10.1016/j.bbrc.2008.09.046, 1:CAS:528:DC%2BD1cXht1ensrfJ
Li X, Wang X, He K, et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell, 2008, 20: 259–276 18263775, 10.1105/tpc.107.056879, 1:CAS:528:DC%2BD1cXkslSitLk%3D
Dorweiler J E, Carey C C, Kubo K M, et al. Mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell, 2000, 12: 2101–2118 11090212, 10.1105/tpc.12.11.2101, 1:CAS:528:DC%2BD3MXos1Gr
Vaughn M W, Tanurd Ić M, Lippman Z, et al. Epigenetic Natural Variation in Arabidopsis thaliana. PLoS Biol, 2007, 5(7): e174, doi:10.1371/journal.pbio.0050174
Finnegan E J, Peacock W J, Dennis E S. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA, 1996, 93: 8449–8454 8710891, 10.1073/pnas.93.16.8449, 1:CAS:528:DyaK28XkvFyksLg%3D
Chan S W, Henderson I R, Zhang X, et al. RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in Arabidopsis. PLoS Genet, 2006, 2: e83. doi:10.1371/journal.p.gen. 0020083
Henderson I R, Jacobsen S E. Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev, 2008, 22: 1597–1606 18559476, 10.1101/gad.1667808, 1:CAS:528:DC%2BD1cXnvVWmsbw%3D
Kakutani T, Jeddeloh J A, Flowers S K, et al. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci USA, 1996, 93: 12406–12411 8901594, 10.1073/pnas.93.22.12406, 1:CAS:528:DyaK28Xms1Gjurc%3D
Saze H, Kakutani T. Heritable epigenetic mutation of a transposon flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J, 2007, 26: 3641–3652 17627280, 10.1038/sj.emboj.7601788, 1:CAS:528:DC%2BD2sXos1eqtb8%3D
Dennis E S, Peacock W J. Epigenetic regulation of flowering. Curr Opin Plant Biol, 2007, 10: 1–8 10.1016/j.pbi.2007.06.009, 1:CAS:528:DC%2BD2sXhtFekurjE
Swiezewski S, Crevillen P, Liu F, et al. Small RNA-mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc Natl Acad Sci USA, 2007, 104: 3633–3638 17360694, 10.1073/pnas.0611459104, 1:CAS:528:DC%2BD2sXjtVWrsrY%3D
Baurle I, Smith L, Baulcombe D C, et al. Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science, 2007, 318: 109–112 17916737, 10.1126/science.1146565, 1:CAS:528:DC%2BD2sXhtFWitb%2FL
Lu C Tej S S, Luo S, et al. Elucidation of the small RNA component of the transcriptome. Science, 2005, 309: 1567–1569 16141074, 10.1126/science.1114112, 1:CAS:528:DC%2BD2MXpsFWju7c%3D
Huh J H, Bauer M J, Hsieh T F, et al. Endosperm gene imprinting and seed development. Curr Opin Genet Dev, 2007, 17: 480–485 17962010, 10.1016/j.gde.2007.08.011, 1:CAS:528:DC%2BD2sXhsVWisb%2FK
Soppe W J, Jacobsen S E, Alonso-Blanco C, et al. The late flowering phenotype of fwa mutants is caused by gain-of function epigenetic alleles of a homeodomain gene. Mol Cell, 2000, 6: 791–802 11090618, 10.1016/S1097-2765(05)00090-0, 1:CAS:528:DC%2BD3cXnvVegt74%3D
Woo H R, Dittmer, T A, Richards E J. Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet, 2008, 4(8): e1000156
Slotkin R K, Vaughn M, Borges F, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell, 2009, 136: 461–472 19203581, 10.1016/j.cell.2008.12.038, 1:CAS:528:DC%2BD1MXltFSnsro%3D
Chinnusamy V, Zhu J K. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol, 2009, 12: 133–139 19179104, 10.1016/j.pbi.2008.12.006, 1:CAS:528:DC%2BD1MXjsVOgtbw%3D
Steward N, Kusano T, Sano H. Expression of ZmMET1, a gene encoding a DNA methyltransferase from maize, is associated not only with DNA replication in actively proliferating cells, but also with altered DNA methylation status in cold-stressed quiescent cells. Nucleic Acids Res, 2000, 28: 3250–3259 10954592, 10.1093/nar/28.17.3250, 1:STN:280:DC%2BD3M%2Fht1Gguw%3D%3D
Hashida S N, Uchiyama T, Martin C, et al. The temperature-dependent change in methylation of the Antirrhinum transposon Tam3 is controlled by the activity of its transposase. Plant Cell, 2006, 18: 104–118 16326924, 10.1105/tpc.105.037655, 1:CAS:528:DC%2BD28XptlGgtQ%3D%3D
Steward N, Ito M, Yamaguchi Y, et al. Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J Biol Chem, 2002, 277: 37741–37746 12124387, 10.1074/jbc.M204050200, 1:CAS:528:DC%2BD38XnsVejtrk%3D
Duan K, Ding X, Zhang Q, et al. AtCopeg1, the unique gene originated from AtCopia95 retrotran- sposon family, is sensitive to external hormones and abiotic stresses. Plant Cell Rep, 2008, 27: 1065–1073 18309491, 10.1007/s00299-008-0520-2, 1:CAS:528:DC%2BD1cXmtFenu7o%3D
Kashkush K, Feldman M, Levy A A. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet, 2003, 33: 102–106 12483211, 10.1038/ng1063, 1:CAS:528:DC%2BD38XpvVeqs7s%3D
Kashkush K, Khasdan V. Large-scale survey of cytosine methylation of retrotransposons and the impact of readout transcription from long terminal repeats on expression of adjacent rice genes. Genetics, 2007, 177: 1975–1985 18073417, 10.1534/genetics.107.080234, 1:CAS:528:DC%2BD1cXhtleqtb0%3D
Choi C S, Sano H. Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol Genet Genome, 2007, 277: 589–600 10.1007/s00438-007-0209-1, 1:CAS:528:DC%2BD2sXksVarsbw%3D
Kovarik A, Koukalova B, Bezdek M, et al. Hypermethylation of tobacco heterochromatic loci in response to osmotic stress. Theor Appl Genet, 1997, 95: 301–306 10.1007/s001220050563
Labra M, Ghiani A, Citterio S, et al. Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant Biol (Stuttgart), 2002, 4: 694–699 10.1055/s-2002-37398, 1:CAS:528:DC%2BD3sXis1elu7s%3D
Dyachenko O V, Zakharchenko N S, Shevchuk T V, et al. Effect of hypermethylation of CCWGG sequences in DNA of Mesembryanthemum crystallinum plants on their adaptation to salt stress. Biochem (Moscow), 2006, 71: 461–465 10.1134/S000629790604016X, 1:CAS:528:DC%2BD28Xjs1KktLw%3D
Sunkar R, Zhu J K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell, 2004, 16: 2001–2019 15258262, 10.1105/tpc.104.022830, 1:CAS:528:DC%2BD2cXmvVansb0%3D
Borsani O, Zhu J H, Verslues P E, et al. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell, 2005, 123: 1279–1291 16377568, 10.1016/j.cell.2005.11.035, 1:CAS:528:DC%2BD28XktlWitQ%3D%3D
Jin H, Vacic V, Girke T, et al. Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol Biol, 2008, 9:6, doi: 10.1186/1471-2199-9-6 18194570, 10.1186/1471-2199-9-6, 1:CAS:528:DC%2BD1cXmt12ksLo%3D
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chinnusamy, V., Zhu, JK. RNA-directed DNA methylation and demethylation in plants. SCI CHINA SER C 52, 331–343 (2009). https://doi.org/10.1007/s11427-009-0052-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-009-0052-1