Abstract
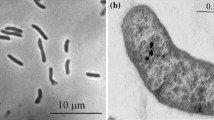
An anaerobic, rod-shaped, mesophilic, chemolithoautotrophic, sulfate-reducing bacterial strain IOR2T was isolated from a newly found deep-sea hydrothermal vent (OVF, Onnuri Vent Field) area in the central Indian Ocean ridge (11°24′88″ S 66°25′42″ E, 2021 m water depth). The 16S rRNA gene sequence analysis revealed that the strain IOR2T was most closely related to Desulfovibrio senegalensis BLaC1T (96.7%). However, it showed low similarity with the members of the family Desulfovibrionaceae, such as Desulfovibrio tunisiensis RB22T (94.0%), D. brasiliensis LVform1T (93.9%), D. halophilus DSM 5663T (93.7%), and Pseudodesulfovibrio aespoeensis Aspo-2T (93.2%). The strain IOR2T could grow at 23–42°C (optimum 37°C), pH 5.0–8.0 (optimum pH 7.0) and with 0.5–6.5% (optimum 3.0%) NaCl. The strain could use lactate, pyruvate, H2, and glycerol as electron donors and sulfate, thiosulfate, and sulfite as electron acceptors. The major fatty acids of the strain IOR2T were iso-C15:0, iso-C17:0, ante-iso-C15:0, and summed feature 9 (C16:0 methyl/iso-C17:1ω9c). Both the strains IOR2T and BLaC1T could grow with CO2 and H2 as the sole sources of carbon and energy, respectively. Genomic evidence for the Wood-Ljungdahl pathway in both the strains reflects chemolithoautotrophic growth. The DNA G + C content of the strain IOR2T and BLaC1T was 58.1–60.5 mol%. Based on the results of the phylogenetic and physiologic studies, Paradesulfovibrio onnuriensis gen. nov., sp. nov. with the type strain IOR2T (= KCTC 15845T = MCCC 1K04559T) was proposed to be a member of the family Desulfovibrionaceae. We have also proposed the reclassification of D. senegalensis as Paradesulfovibrio senegalensis comb. nov.
Similar content being viewed by others
References
Bale, S.J., Goodman, K., Rochelle, P.A., Marchesi, J.R., Fry, J.C., Weightman, A.J., and Parkes, R.J. 1997. Desulfovibrio profundus sp. nov., a novel barophilic sulfate-reducing bacterium from deep sediment layers in the Japan Sea. Int. J. Syst. Bacteriol.47, 515–521.
Ben Ali Gam, Z., Oueslati, R., Abdelkafi, S., Casalot, L., Tholozan, J.L., and Labat, M. 2009. Desulfovibrio tunisiensis sp. nov., a novel weakly halotolerant, sulfate-reducing bacterium isolated from exhaust water of a Tunisian oil refinery. Int. J. Syst. Evol. Microbiol.59, 1059–1063.
Cao, J., Gayet, N., Zeng, X., Shao, Z., Jebbar, M., and Alain, K. 2016. Pseudodesulfovibrio indicus gen. nov., sp. nov., a piezophilic sulfate-reducing bacterium from the Indian Ocean and reclassification of four species of the genus Desulfovibrio. Int. J. Syst. Evol. Microbiol.66, 3904–3911.
Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol.17, 368–376.
Finster, K.W. and Kjeldsen, K.U. 2010. Desulfovibrio oceani subsp. oceani sp. nov., subsp. nov. and Desulfovibrio oceani subsp. galateae subsp. nov., novel sulfate-reducing bacteria isolated from the oxygen minimum zone off the coast of Peru. Antonie van Leeuwenhoek97, 221–229.
Fitch, W.M. 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool.20, 406–416.
Gibson, G.R. 1990. Physiology and ecology of the sulphate-reducing bacteria. J. Appl. Bacteriol.69, 769–797.
Giovannoni, S.J. 1991. The polymerase chain reaction, In Stackebrandt, E. and Goodfellow, M. (eds.), Nucleic acid techniques in bacterial systematics. 1st edn, pp. 177–203. John Wiley & Sons Ltd., Chichester, England.
Hamdi, O., Ben Hania, W., Postec, A., Bartoli, M., Hamdi, M., Bouallagui, H., Fauque, G., Ollivier, B., and Fardeau, M.L. 2013. Isolation and characterization of Desulfocurvus thunnarius sp. nov., a sulfate-reducing bacterium isolated from an anaerobic sequencing batch reactor treating cooking wastewater. Int. J. Syst. Evol. Microbiol.63, 4237–4242.
Heidelberg, J.F., Seshadri, R., Haveman, S.A., Hemme, C.L., Paulsen, I.T., Kolonay, J.F., Eisen, J.A., Ward, N., Methe, B., Brinkac, L.M., et al. 2004. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol.22, 554–559.
Jukes, T.H. and Cantor, C.R. 1969. Evolution of protein molecules, In Munro, H.N. and Allison, J.B. (eds.) Mammalian protein metabolism, vol. 3. pp. 21–132. Academic Press, New York, USA.
Khelaifia, S., Fardeau, M.L., Pradel, N., Aussignargues, C., Garel, M., Tamburini, C., Cayol, J.L., Gaudron, S., Gaill, F., and Ollivier, B. 2011. Desulfovibrio piezophilus sp. nov., a piezophilic, sulfatereducing bacterium isolated from wood falls in the Mediterranean Sea. Int. J. Syst. Evol. Microbiol.61, 2706–2711.
Kotsyurbenko, O.R., Simankova, M.V., Nozhevnikova, A.N., Zhilina, T.N., Bolotina, N.P., Lysenko, A.M., and Osipov, G.A. 1995. New species of psychrophilic acetogens: Acetobacterium bakii sp. nov., A. paludosum sp. nov., A. fimetarium sp. nov. Arch. Microbiol.163, 29–34.
Kuever, J., Rainey, F.A., and Widdel, F. 2005. Genus I. Desulfovibrio Kluyver and van Niel 1936, 397AL. In Brenner, D.J., Krieg, N.R., and Staley, J.T. (eds.), Bergey’s manual of systematic bacteriology, vol. 2, pp. 926–938. Springer, New York, USA.
Matias, P.M., Pereira, I.A., Soares, C.M., and Carrondo, M.A. 2005. Sulphate respiration from hydrogen in Desulfovibrio bacteria: a structural biology overview. Prog. Biophys. Mol. Biol.89, 292–329.
Morais-Silva, F.O., Rezende, A.M., Pimentel, C., Santos, C.I., Clemente, C., Varela-Raposo, A., Resende, D.M., da Silva, S.M., de Oliveira, L.M., Matos, M., et al. 2014. Genome sequence of the model sulfate reducer Desulfovibrio gigas: a comparative analysis within the Desulfovibrio genus. Microbiologyopen3, 513–530.
Motamedi, M. and Pedersen, K. 1998. Desulfovibrio aespoeensis sp. nov., a mesophilic sulfate-reducing bacterium from deep ground-water at Äspö hard rock laboratory, Sweden. Int. J. Syst. Bacteriol.48, 311–315.
Na, S.I., Kim, Y.O., Yoon, S.H., Ha, S.M., Baek, I., and Chun, J. 2018. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol.56, 280–285.
Nakazawa, H., Arakaki, A., Narita-Yamada, S., Yashiro, I., Jinno, K., Aoki, N., Tsuruyama, A., Okamura, Y., Tanikawa, S., Fujita, N., et al. 2009. Whole genome sequence of Desulfovibrio magneticus strain RS-1 revealed common gene clusters in magnetotactic bacteria. Genome. Res.19, 1801–1808.
Postgate, J.R. and Campbell, L.L. 1966. Classification of Desulfovibrio species, the nonsporulating sulfate-reducing bacteria. Bacteriol. Rev.30, 732–738.
Ranchou-Peyruse, M., Goñi-Urriza, M., Guignard, M., Goas, M., Ranchou-Peyruse, A., and Guyoneaud, R. 2018. Pseudodesulfovibrio hydrargyri sp. nov., a mercury-methylating bacterium isolated from a brackish sediment. Int. J. Syst. Evol. Microbiol.68, 1461–1466.
Robb, F.T., Place, A.R., Sowers, K.R., Schreier, H.J., DasSarma, S., and Fleischmann, E.M. 1995. In Robb, F.T., Sowers, K.R., Place, A.R., Schreier, H.J. Archaea: a laboratory manual, pp. 3–29. Cold spring harbor laboratory press, New York, USA.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol.4, 406–425.
Sass, H., Ramamoorthy, S., Yarwood, C., Langner, H., Schumann, P., Kroppenstedt, R.M., Spring, S., and Rosenzweig, R.F. 2009. Desulfovibrio idahonensis sp. nov., sulfate-reducing bacteria isolated from a metal (loid)-contaminated freshwater sediment. Int. J. Syst. Evol. Microbiol.59, 2208–2214.
Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. MIDI Inc, Newark, DE, USA.
Sato, T., Fukui, T., Atomi, H., and Imanaka, T. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol.185, 210–220.
Sheik, C.S., Sieber, J.R., Badalamenti, J.P., Carden, K., and Olson, A. 2017. Complete genome sequence of Desulfovibrio desulfuricans strain G11, a model sulfate-reducing, hydrogenotrophic, and syntrophic partner organism. Genome Announc.5, e01207–17.
Shivani, Y., Subhash, Y., Sasikala, C., and Ramana, C.V. 2017. Halodesulfovibrio spirochaetisodalis gen. nov. sp. nov. and reclassification of four Desulfovibrio spp. Int. J. Syst. Evol. Microbiol.67, 87–93.
Suzuki, D., Ueki, A., Amaishi, A., and Ueki, K. 2009. Desulfovibrio portus sp. nov., a novel sulfate-reducing bacterium in the class Deltaproteobacteria isolated from an estuarine sediment. J. Gen. Appl. Microbiol.55, 125–133.
Takii, S., Hanada, S., Hase, Y., Tamaki, H., Uyeno, Y., Sekiguchi, Y., and Matsuura, K. 2008. Desulfovibrio marinisediminis sp. nov., a novel sulfate-reducing bacterium isolated from coastal marine sediment via enrichment with casamino acids. Int. J. Syst. Evol. Microbiol.58, 2433–2438.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol.28, 2731–2739.
Tatusova, T., Dicuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E.P., Zaslavsky, L., Lomsadze, A., Pruitt, K.D., Borodovsky, M., and Ostell, J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res.44, 6614–6624.
Thabet, O.B., Wafa, T., Eltaief, K., Cayol, J.L., Hamdi, M., Fauque, G., and Fardeau, M.L. 2011. Desulfovibrio legallis sp. nov.: A moderately halophilic, sulfate-reducing bacterium isolated from a wastewater digestor in Tunisia. Curr. Microbiol.62, 486–491.
Thioye, A., Gam, Z.B.A., Mbengue, M., Cayol, J.L., Joseph-Bartoli, M., Touré-Kane, C., and Labat, M. 2017. Desulfovibrio senegalensis sp. Nov., a mesophilic sulfate reducer isolated from marine sediment. Int. J. Syst. Evol. Microbiol.67, 3162–3166.
van der Hoeven, J.S., van den Kieboom, C.W., and Schaeken, M.J. 1995. Sulfate-reducing bacteria in the periodontal pocket. Oral Microbiol. Immunol.10, 288–290.
Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F.O., Ludwig, W., Schleifer, K.H., Whitman, W.B., Euzéby, J., Amann, R., and Rosselló-Móra, R. 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol.12, 635–645.
Yoon, S.H., Ha, S.M., Kwon, S., Lim, J., Kim, Y., Seo, H., and Chun, J. 2017a. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol.67, 1613–1617.
Yoon, S.H., Ha, S.M., Lim, J., Kwon, S., and Chun, J. 2017b. A largescale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek110, 1281–1286.
Zhao, C., Gao, Z., Qin, Q., Li, F., and Ruan, L. 2012. Desulfobaculum xiamenensis gen. nov., sp. nov., a member of the family Desulfovibrionaceae isolated from marine mangrove sediment. Int. J. Syst. Evol. Microbiol.62, 1570–1575.
Acknowledgments
This work was supported by the KIOST In-house Program (PE99822) and the understanding of the deep-sea biosphere on seafloor hydrothermal vents in the Indian Ridge Program (No. 20170411) of the Ministry of Ocean and Fisheries of the Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors have no conflicts of interest to declare.
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kim, Y.J., Yang, JA., Lim, J.K. et al. Paradesulfovibrio onnuriensis gen. nov., sp. nov., a chemolithoautotrophic sulfate-reducing bacterium isolated from the Onnuri vent field of the Indian Ocean and reclassification of Desulfovibrio senegalensis as Paradesulfovibrio senegalensis comb. nov.. J Microbiol. 58, 252–259 (2020). https://doi.org/10.1007/s12275-020-9376-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-020-9376-0