Abstract
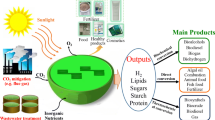
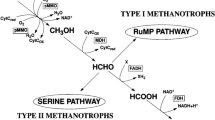
Fueled by the recognition of hydrogen as a promising renewable energy source for the future, there have been many attempts to find greener and more economical ways for its production from various sources. In this study, Methylomonas sp. DH-1, a type I methanotroph, was found to produce hydrogen using methane as a sole carbon source, under micro-aerobic conditions; this is analogous to the partial oxidation of methane in a thermochemical process based on metal catalysts. Flask cultures of Methylomonas sp. DH-1 were used to investigate the effects of different culture conditions on hydrogen production, including oxygen levels, methane/oxygen ratios, and initial cell densities. Methylomonas sp. DH-1 could produce hydrogen at an oxygen level below 4%, regardless of the methane content in the flask, implying that the critical factor for hydrogen production is the oxygen level, rather than the methane/oxygen ratio. Moreover, Methylomonas sp. DH-1 shows reversibility in hydrogen production and uptake, because the strain produces hydrogen under micro-aerobic conditions, uptakes it when the oxygen levels increase, and restores the hydrogen production capability when conditions become microaerobic again. Under initial conditions of 30% methane, 70% air, and an OD600nm of 6, hydrogen production was 26.87 μmol and its yields per methane and dry cell weight were 14.98 mmol-H2/mol-CH4 and 101.53 μmol-H2/g DCW, respectively, after 24 h of cultivation.
Similar content being viewed by others
References
Ergal, İ., W. Fuchs, B. Hasibar, B. Thallinger, G. Bochmann, and S. K. M. R. Rittmann (2018) The physiology and biotechnology of dark fermentative biohydrogen production. Biotechnol. Adv. 36: 2165–2186.
Maintinguer, S. I., C. Z. Lazaro, R. Pachiega, M. B. A. Varesche, R. Sequinel, and J. E. de Oliveira (2017) Hydrogen bioproduction with Enterobacter sp. isolated from brewery wastewater. Int. J. Hydrog. Energy. 42: 152–160.
Kim, M. S., H. H. Kim, K. M. Lee, H. J. Lee, and C. Lee (2017) Oxidation of microcystin-LR by ferrous-tetrapolyphosphate in the presence of oxygen and hydrogen peroxide. Water Res. 114: 277–3.
Liu, G. and J. Shen (2004) Effects of culture and medium conditions on hydrogen production from starch using anaerobic bacteria. J. Biosci. Bioeng. 98: 251–3.
Loipersböck, J., M. Lenzi, R. Rauch, and H. Hofbauer (2017) Hydrogen production from biomass: The behavior of impurities over a CO shift unit and a biodiesel scrubber used as a gas treatment stage. Korean J. Chem. Eng. 34: 2198–3.
Kidanu, W. G., P. T. Trang, and H. H. Yoon (2017) Hydrogen and volatile fatty acids production from marine macroalgae by anaerobic fermentation. Biotechnol. Bioprocess Eng. 22: 612–3.
Laurinavichene, T. V., N. A. Zorin, and A. A. Tsygankov (2002) Effect of redox potential on activity of hydrogenase 1 and hydrogenase 2 in Escherichia coli. Arch. Microbiol. 178: 437–3.
Cracknell, J. A., A. F. Wait, O. Lenz, B. Friedrich, and F. A. Armstrong (2009) A kinetic and thermodynamic understanding of O2 tolerance in [NiFe]-hydrogenases. Proc. Natl. Acad. Sci. USA. 106: 20681–3.
Murphy, B. J., F. Sargent, and F. A. Armstrong (2014) Transforming an oxygen-tolerant [NiFe] uptake hydrogenase into a proficient, reversible hydrogen producer. Energy Envrion. Sci. 7: 1426–3.
Hwang, I. Y., D. H. Hur, J. H. Lee, C. H. Park, I. S. Chang, J. W. Lee, and E. Y. Lee (2015) Batch conversion of methane to methanol using Methylosinus trichosporium OB3b as biocatalyst. J. Microbiol. Biotechnol. 25: 375–3.
Haynes, C. A. and R. Gonzalez (2014) Rethinking biological activation of methane and conversion to liquid fuels. Nat. Chem. Biol. 10: 331–3.
Hong, E., S. A. Jeon, S. S. Lee, and C. H. Shin (2018) Methane combustion over Pd/Ni-Al oxide catalysts: Effect of Ni/Al ratio in the Ni-Al oxide support. Korean J. Chem. Eng. 35: 1815–3.
Hanson, R. S. and T. E. Hanson (1996) Methanotrophic bacteria. Microbiol. Rev. 60: 439–3.
Fei, Q., M. T. Guarnieri, L. Tao, L. M. Laurens, N. Dowe, and P. T. Pienkos (2014) Bioconversion of natural gas to liquid fuel: opportunities and challenges. Biotechnol. Adv. 32: 596–3.
Kalyuzhnaya, M. G., A. W. Puri, and M. E. Lidstrom (2015) Metabolic engineering in methanotrophic bacteria. Metab. Eng. 29: 142–3.
Lee, O. K., D. H. Hur, D. T. Nguyen, and E. Y. Lee (2016) Metabolic enginee. Biofuel Bioprod. Biorefin. 10: 848–3.
Kalyuzhnaya, M. G., S. Yang, O. N. Rozova, N. E. Smalley, J. Clubb, A. Lamb, G. A. Nagana Gowda, D. Raftery, Y. Fu, F. Bringel, S. Vuilleumier, D. A. C. Beck, Y. A. Trotsenko, V. N. Khmelenina, and M. E. Lidstrom (2013) Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat. Commun. 4: 2785.
Gilman, A., Y. Fu, M. Hendershott, F. Chu, A. W. Puri, A. L. Smith, M. Pesesky, R. Lieberman, D. A. C. Beck, and M. E. Lidstrom (2017) Oxygen-limited metabolism in the methanotroph Methylomicrobium buryatense 5GB1C. Peer J. 5: e3945.
Hur, D. H., J. G. Na, and E. Y. Lee (2017) Highly efficient bioconversion of methane to methanol using a novel type I Methylomonas sp. DH-1 newly isolated from brewery waste sludge. J. Chem. Technol. Biotechnol. 92: 311–3.
Nguyen, A. D., D. Kim, and E. Y. Lee (2019) A comparative transcriptome analysis of the novel obligate methanotroph Methylomonas sp. DH-1 reveals key differences in transcriptional responses in C1 and secondary metabolite pathways during growth on methane and methanol. BMC Genomics. 20: 130.
Burgdorf, T., O. Lenz, T. Buhrke, E. Van Der Linden, A. K. Jones, S. P. Albracht, and B. Friedrich (2005) [NiFe]-hydrogenases of Ralstonia eutropha H16: modular enzymes for oxygentolerant biological hydrogen oxidation. J. Mol. Microbiol. Biotechnol. 10: 181–3.
Nguyen, A. D., I. Y. Hwang, O. K. Lee, D. H. Hur, Y. C. Jeon, S. Hadiyati, M. S. Kim, S. H. Yoon, H. Jeong, and E. Y. Lee (2018) Functional analysis of Methylomonas sp. DH-1 genome as a promising biocatalyst for bioconversion of methane to valuable chemicals. Catalysts. 8: 117–3.
Baritugo, K. A., H. T. Kim, Y. C. David, J. H. Choi, J. Choi, T. W. Kim, C. Park, S. H. Hong, J. G. Na, K. J. Jeong, J. C. Joo, and S. J. Park (2018) Recent advances in metabolic engineering of Corynebacterium glutamicum strains as potential platform microorganisms for biorefinery. Biofuel Bioprod. Bior. 12: 899–3.
Baritugo, K. A., H. T. Kim, Y. David, J. Choi, S. H. Hong, K. J. Jeong, J. H. Choi, J. C. Joo, and S. J. Park (2018) Metabolic engineering of Corynebacterium glutamicum for fermentative production of chemicals in biorefinery. Appl. Microbiol. Biotechnol. 102: 3915–3.
David, Y., M. G. Baylon, S. D. V. N. Pamidimarri, K. A. Baritugo, C. G. Chae, Y. J. Kim, T. W. Kim, M. S. Kim, J. G. Na, and S. J. Park (2017) Screening of microorganisms able to degrade low-rank coal in aerobic conditions: Potential coal biosolubilization mediators from coal to biochemicals. Biotechnol. Bioprocess Eng. 22: 178–3.
Jeon, H. S., S. E. Park, B. Ahn, and Y. K. Kim (2017) Enhancement of biodiesel production in Chlorella vulgaris cultivation using silica nanoparticles. Biotechnol. Bioprocess Eng. 22: 136–3.
Oh, Y. H., I. Y. Eom, J. C. Joo, J. H. Yu, B. K. Song, S. H. Lee, S. H. Hong, and S. J. Park (2015) Recent advances in development of biomass pretreatment technologies used in biorefinery for the production of bio-based fuels, chemicals and polymers. Korean J. Chem. Eng. 32: 1945–3.
Acknowledgements
This research was supported by the Sogang University Research Grant of 2017 (201710069.01) and by “Human Resources Program in Energy Technology” of the Korea Institute of Energy Technology Evaluation and Planning (KETEP), granted financial resources from the Ministry of Trade, Industry & Energy, Republic of Korea (No. 20174010201150).
The authors declare no conflict of interest.
Neither ethical approval nor informed consent was required for this study.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Materials
Rights and permissions
About this article
Cite this article
Jo, S.Y., Rhie, M.N., Jung, S.M. et al. Hydrogen Production from Methane by Methylomonas sp. DH-1 under Micro-aerobic Conditions. Biotechnol Bioproc E 25, 71–77 (2020). https://doi.org/10.1007/s12257-019-0256-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-019-0256-6