Abstract
Environmental DNA (eDNA) detection methods can complement traditional biomonitoring to yield new ecological insights in aquatic systems. However, the conceptual and methodological frameworks for aquatic eDNA detection and interpretation were developed primarily in freshwater environments and have not been well established for estuaries and marine environments that are by nature dynamic, turbid, and hydrologically complex. Environmental context and species life history are critical for successful application of eDNA methods, and the challenges associated with eDNA detection in estuaries were the subject of a symposium held at the University of California Davis on January 29, 2020 (https://marinescience.ucdavis.edu/engagement/past-events/edna). Here, we elaborate upon topics addressed in the symposium to evaluate eDNA methods in the context of monitoring and biodiversity studies in estuaries. We first provide a concise overview of eDNA science and methods, and then examine the San Francisco Estuary (SFE) as a case study to illustrate how eDNA detection can complement traditional monitoring programs and provide regional guidance on future potential eDNA applications. Additionally, we offer recommendations for enhancing communication between eDNA scientists and natural resource managers, which is essential for integrating eDNA methods into existing monitoring programs. Our intent is to create a resource that is accessible to those outside the field of eDNA, especially managers, without oversimplifying the challenges or advantages of these methods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Environmental DNA (eDNA) capture methods provide organism detection data that can inform critical management decisions (Jerde 2019). There is a growing potential for information derived from eDNA detections of organisms to be generated more extensively and used in decision making in estuarine systems, many of which are in need of new monitoring tools to help address difficult management challenges (Cloern et al. 2016a). Here, we focus primarily on eDNA isolated from water, but we note that eDNA methods also may be useful in analysis of soil and aquatic sediments (e.g., Turner et al. 2015), which in estuaries includes exposed tidal areas and benthic habitats.
This paper is intended for both eDNA scientists and non-specialists, including managers who might be considering eDNA methods in their monitoring programs. Because an accurate understanding of what eDNA is and how it can be detected is essential for evaluating its potential usefulness in management (Hansen et al. 2018; Sepulveda et al. 2020c), we first provide a primer on the basics of eDNA science and methodology. Following this foundational knowledge, we investigate the potential for eDNA methods to support estuarine management. The San Francisco Estuary (SFE) is used as an example for discussion of the considerations and challenges of eDNA methods although each estuary is unique and should be considered independently. Finally, because we have observed that misconceptions and lack of communication between eDNA scientists and managers can hinder accurate understanding of eDNA methods, we make general recommendations to improve communication between these two groups.
A Brief Guide to eDNA Science and Methods
“Environmental DNA” is not a method; it is mixture of DNA molecules that can be detected by multiple, continually evolving technologies (Lacoursière‐Roussel and Deiner 2019). DNA molecules that are shed, excreted, or otherwise derived from organisms and left behind in the environment comprise eDNA. The environment can be water, soil, air, and the DNA can originate from living or dead organisms. Here, we mostly focus on detection of extra-organismal DNA shed from species, but we note recent discussions in the literature re-emphasizing that “eDNA” includes all DNA in an environmental sample, including that derived from whole microorganisms as well as microscopic life stages of macroorganisms (e.g., veliger larvae of mollusks) (Pawlowski et al. 2020, 2021; Rodriguez-Ezpeleta et al. 2021). This distinction is more than just semantic, as interpreting species presence indirectly via extra-organismal eDNA detection presents more challenges than direct detection from whole and sampled individuals (Rodriguez-Ezpeleta et al. 2021).
Environmental DNA detection methods are divided into those detecting single or a small number of species, and those focused on whole communities (environmental metabarcoding or metagenomics). The origins and mechanics of eDNA, and how to most efficiently and accurately detect it, remain areas of active research due to the dynamic effects of biological and environmental processes. Overall, the field is developing an understanding of when and how protocols can be standardized and when specialized approaches are needed. Several recent reviews have provided authoritative guidance on broad conceptual and methodological topics related to eDNA (Barnes and Turner 2016; Goldberg et al. 2016; Tsuji et al. 2019; Deiner et al. 2021); see Table 1 for recommended review papers and online eDNA resources. Here, we summarize the most important aspects of eDNA science and method development as they relate to estuaries. We define estuaries as marine, brackish, and freshwater environments subject to both oceanic and riverine influence, with the acknowledgment that estuaries vary enormously in terms of size, hydrogeography, anthropomorphic influence, and other important characteristics.
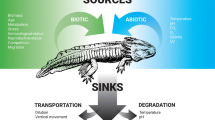
Environmental DNA Origins and Dynamics
Macroorganisms release eDNA into their surroundings (Ficetola et al. 2008) primarily through feces and urine, epithelial cells in external mucus, sloughed scales, and gametes (Barnes and Turner 2016; Stewart 2019). This type of eDNA co-occurs in environmental samples along with intact microorganisms (containing their own DNA), for example phytoplankton and zooplankton in aquatic samples. Since the size and physical properties of the cells or particles impact the physical properties of the eDNA, like transport and degradation, data on eDNA particle size provide important foundational information. For microorganismal eDNA, the cell sizes are generally known a priori. In contrast, aquatic eDNA from macroscopic aquatic organisms exists in several possible physical states: multicellular pieces of tissue, single cells, organelles, free extracellular DNA, or any of these bound to other particles. To date, there are very limited published data on macroorganismal eDNA particle size ranges. The size distribution of particles containing the most abundant fish eDNA in two freshwater studies ranged between 1 and 10 µm (Turner et al. 2014; Wilcox et al. 2015). In contrast, eDNA concentrations were highest in the 0- to 0.2-µm particle size range for an aquatic invertebrate (Moushomi et al. 2019). Environmental conditions, including chlorophyll and turbidity, can affect the observed eDNA particle size distribution (Barnes et al. 2021). Similarly, the rate of eDNA shedding is context dependent and can be affected by both biotic and abiotic factors; for example, the life history, biomass, or density of the target organism and water temperature (Klymus et al. 2015; Lacoursière-Roussel et al. 2016; Sassoubre et al. 2016). In addition to shedding rate, the amount of eDNA at a specific location is dependent on degradation, which reduces eDNA concentrations, and transport, which can both reduce eDNA at its source and concentrate it away from its source (Andruszkiewicz et al. 2020; Thalinger et al. 2020b).
Environmental DNA can be transported from its point of origin by diffusion and advection. Its transport dynamics differ from conservative tracers (substances used to track water flow that do not degrade or change) and are more consistent with heterogenous particles that vary in size, degrade over time, and can also bind to substrates (Jerde et al. 2016; Fremier et al. 2019). In freshwater lotic systems, differences in hydrology and residence time of organisms result in maximum detection distances ranging from < 5 m to > 100 km (Deiner and Altermatt 2014; Pilliod et al. 2014; Jane et al. 2015; Shogren et al. 2017; Pont et al. 2018). Despite this wide range, eDNA transport in these systems can be modeled effectively and is perhaps surprisingly predictable. First, modeling eDNA as fine particulate organic matter (FPOM) and integrating location-specific hydrological information allows accurate prediction of the distance of detection in rivers, with deep and fast rivers transporting eDNA farther than shallower ones (Pont et al. 2018). Second, using a bidirectional hydrodynamic model that includes both downstream advection as well as lateral mixing can work well in a large river, explaining positive detections directly downstream of an eDNA source but negligible detections laterally near the source (not directly downstream) (Laporte et al. 2020). There are fewer studies on transport in marine systems, but there are data supporting both limited dispersal in nearshore environments (Kelly et al. 2018) and eDNA transport up to 4 km from its source (Baker et al. 2018). Marine particle tracking models predict that dispersal is possible up to tens of kilometers (Andruszkiewicz et al. 2019). In low-flow marine conditions, low levels of mixing can create a steep eDNA concentration gradient decreasing away from the source (Eble et al. 2020). For example, in a kelp forest, different species assemblages at habitats separated by as little as 60 m could be effectively discriminated (Port et al. 2016). These findings mirror data from freshwater lentic systems, where limited detection at > 40 m has been observed (Dunker et al. 2016; Goldberg et al. 2018).
Similar to eDNA shedding, eDNA degradation is impacted by biotic and abiotic factors including eDNA state, temperature, binding to substrate biofilms, and the length and conformation of the DNA molecules (Barnes et al. 2014; Strickler et al. 2015; Eichmiller et al. 2016; Shogren et al. 2018; Jo et al. 2019). Generally, eDNA degrades more slowly in cold, alkaline aquatic conditions with low solar ultraviolet B (UV-B) radiation (Pilliod et al. 2014; Strickler et al. 2015). Microbial-mediated eDNA degradation and extracellular enzymes like nucleases are also important for decay dynamics; these can vary between habitats and are related to abiotic factors like temperature (Barnes et al. 2014; Harrison et al. 2019). Environmental DNA decay in marine and brackish water has shown similar rates compared to freshwater, although persistence times seem to be lower for unknown reasons (Hansen et al. 2018).
Environmental DNA Capture and Analysis
The collection of aquatic eDNA samples can be relatively straightforward but requires optimization. Several published guides describe best practices for eDNA studies (e.g., Goldberg et al. 2016; Kumar et al. 2020). Environmental DNA must be concentrated from water because it is usually too dilute for in situ detection. Filtration is a common method for concentrating eDNA. Precipitation and centrifugation are also used; flocculation, a method for coagulating particles, has recently been described (Schill 2020). Filtration is performed in the field or laboratory, usually within 24 h of sample collection to reduce the amount of eDNA degradation. Replicate samples are used to characterize variability, increase detection, and improve occupancy and detection rate estimates. Many crucial elements of eDNA field sampling (e.g., filter type) are best selected for specific applications (Rees et al. 2014; Mize et al. 2019); standardization within a project is crucial but between projects different field sampling approaches, optimized to local conditions, may be needed. These approaches should be determined based on spatial and temporal ecology of the target species, environmental conditions, logistical constraints, and project goals. Ideally, the sampling design maximizes the probability of detection, minimizes the associated time and cost, and reduces the probability of sample contamination to near zero.
After sample collection, the eDNA must be extracted from the sample and targeted genetic regions or “barcodes” are amplified. DNA extraction should be performed in a dedicated eDNA space with appropriate contamination control measures (Goldberg et al. 2016). Next, short single strands of DNA (primers) designed to match DNA from the target taxa are used in polymerase chain reaction (PCR) to create millions of copies of a discrete region of DNA (barcode). Molecular detection methods diverge into two types: targeted single-taxon detection and metabarcoding. Targeted detection often focuses on a single species while multiplex reactions simultaneously detecting several species (Brandl et al. 2015) or detecting a broader taxonomic group without distinguishing species are also possible. Metabarcoding has the potential to detect and identify a large diversity of taxa in parallel (often to species level) across a taxonomic group such as fish (Miya et al. 2015) or many groups within a domain (Leray et al. 2013).
There are multiple molecular methods for targeted detection. The most frequently used is probe-based quantitative PCR (qPCR), in which a specific target DNA sequence is amplified and detected with a fluorescent indicator. If a positive control standard curve is used, qPCR can provide estimation of the absolute quantity of the targeted DNA sequence. As a molecular method, qPCR is well established, robust, reliable, and is used in a wide range of applications from medical diagnostics to food safety. Best practices are well documented (Bustin et al. 2009; Forootan et al. 2017) and are being adapted for eDNA (Klymus et al. 2020; Lesperance et al. 2021). Empirical determination of assay limits is recommended for complete assay validation (Klymus et al. 2020; Thalinger et al. 2020a). In some cases, rapid detection of eDNA is possible in the field using a portable qPCR analysis platform (Sepulveda et al. 2018; Thomas et al. 2020). Another quantitative method, digital PCR (dPCR), separates a PCR reaction into many partitions, with each partition undergoing amplification independently, allowing accurate estimation of DNA abundance even at low concentrations. In direct comparisons, dPCR has consistently outperformed qPCR in terms of sensitivity and accuracy of quantification (Doi et al. 2015; Jerde et al. 2016; Hunter et al. 2017; Mauvisseau et al. 2019; Wood et al. 2019). New methodological innovations are now appearing regularly, including the application of CRISPR enzyme-based assays (Williams et al. 2021) and methods for detection of environmental RNA (eRNA), which might increase spatiotemporal accuracy and discriminate between the presence of living vs. dead organisms (Pochon et al. 2017; Cristescu 2019; von Ammon et al. 2019) as well as encoding other genetic and ecological data with great potential, beyond taxa identification (Tsuri et al. 2021; Yates et al. 2021). Furthermore, eRNA data, in conjunction with host specific immune system responses, could potentially indicate disease status before histopathological effects are observed (Connon et al. 2012; Mordecai et al. 2019; Teffer et al. 2019, 2020).
In contrast to the targeted methods described above, metabarcoding uses high-throughput DNA sequencing to detect hundreds to thousands of taxa from a single sample. There are several important differences to note between these two general approaches. For metabarcoding, sample preparation, sequencing, and bioinformatic data analysis can take weeks to months compared with hours for qPCR. However, unlike targeted detection, metabarcoding can detect species which may not be known from an area, an important consideration for invasive species monitoring. When benchmarked to traditional survey methods, metabarcoding has repeatedly demonstrated good concordance, although there are method-specific biases to consider, as exist for every gear type used for traditional surveys (Kelly et al. 2019). For fish assemblages of less than 100 species, estimates of species richness using metabarcoding are consistently similar or better than traditional survey methods (McElroy et al. 2020).
One key factor currently limiting the broader application of metabarcoding is the patchy availability of high-quality reference sequences for many organisms. Metabarcoding analysis uses reference genetic sequences (barcodes) matched to both organism and gene. Incomplete databases or reliance on barcodes with insufficient genetic diversity can lead to errors in taxa assignment. Curated and comprehensive genetic reference sequence databases are time consuming to compile but allow for species-level taxonomic resolution and are generally more accurate (Schenekar et al. 2020; Stoeckle et al. 2020). Broad taxonomic assignments can be made using phylogenetic methods to identify “features” in a community without requiring species identification. For detecting multiple species across a range of taxa, sequencing multiple different barcodes can provide a robust assessment of the full array of species present (Stat et al. 2017) and can map total community similarity over space, which allows reassessment of habitat classifications and identification of ecotones (Lin et al. 2021).
Both targeted detection and metabarcoding can provide quantitative estimates of the starting number of eDNA molecules derived from a specific macroorganism in an environmental sample, and a growing literature indicates that eDNA-derived estimates of organism abundance and biomass may be possible under some circumstances (Rourke et al. 2022; Spear et al. 2021). However, more refinement is needed before this approach is ready for field application and accuracy may vary widely between species, in different habitats, and between assays (Sepulveda et al. 2021); methods should be tested rigorously in the natural environment in addition to mesocosm experiments (Yates et al. 2019). Metabarcoding has generally been understood to be semi-quantitative for abundance or biomass (Taberlet et al. 2018), but many studies are increasingly showing quantitative results once methods have been optimized (e.g., Kelly et al. 2019). Metabarcoding methods optimization using mock communities of known DNA mixtures, mesocosm studies, and new bioinformatic approaches can decrease or correct for biases introduced by the molecular workflow (e.g., primer bias) (Collins et al. 2019; Kelly et al. 2019; McLaren et al. 2019).
Data interpretation
Interpretation of indirect detections and non-detections as are obtained using eDNA methods is necessarily more nuanced compared with direct detection (Darling and Mahon 2011). Inference potential is directly related to sampling design and how methods are administered. Positive detections can result from the organism of interest present nearby at the time of sampling, or from other, more indirect sources (e.g., predator waste, upstream habitats, wastewater effluent, fish transported by commercial or recreational fishers). A sample-level false-positive detection (“false-positive test”) should be interpreted and reported differently from a “presumed positive site,” i.e., when a positive eDNA detection is not confirmed by a non-eDNA detection (Darling et al. 2021). The rate of false-positive tests can be nearly eliminated by following rigorous cleanliness guidelines and collection and analysis of quality assurance-quality control (QA-QC) samples in both the field and laboratory (Goldberg et al. 2016; Sepulveda et al. 2020a). For presumed positive sites lacking non-eDNA detection confirmation, the potential mechanisms leading to the discrepancy (e.g., horizontal eDNA transport) should be investigated and communicated clearly to managers and non-eDNA experts. As with other methods, non-detection does not prove the target taxon is absent. False-negative detections can be decreased through appropriate replication at several levels, including spatial, temporal, and by including sufficient field sample and PCR replicates (see Rees et al. 2014). Comprehensive assay optimization, taking into account the target organism and field conditions, can also reduce the false-negative rate. Therefore, non-detections should generally be interpreted with caution until methods have been tested under a range of conditions (Mahon et al. 2013).
Environmental DNA studies can make use of modeling to estimate species occurrence. Modeling can be used to account for uncertainty (false negatives) bounded within a given statistical error. For example, in occupancy modeling, detections of target organisms are identified as a biased estimate of the true count (Royle et al. 2005). Each survey is classified as detection or non-detection of the target organism and the model accounts for imperfect detection (i.e., false negatives) that are inherent in all survey methods. For eDNA, both sampling and laboratory processes contribute to imperfect detection and can be modeled using a multilevel occupancy model (Schmidt et al. 2013; Hunter et al. 2015; Dorazio and Erickson 2018). Although most models have focused more on false negatives, there are also published occupancy models more carefully assessing false-positive errors (sample-level false-positive detections, or “false-positive tests,” as discussed above) (Tingley et al. 2021). Environmental DNA occupancy models and other modeling approaches can in turn be used to inform sampling design and improve detection probability. For example, detection probability by distance can be estimated using enclosure studies (Schumer et al. 2019a). Models that directly estimate the effect of predictors (e.g., environmental factors like salinity or experimental differences like sampling volume) on eDNA detection can improve data analysis and inform experimental design. The artemis R package (https://fishsciences.github.io/artemis/index.html) is one such model, developed and tested in the SFE specifically for eDNA qPCR requirements (Espe et al. 2022). The analytical framework accommodates biases such as those that arise from qPCR “censored” data, when the amount of detected eDNA occurs at or below the threshold of detection for an assay (Espe et al. 2022). Purposefully designing studies under realistic circumstances is particularly important for management applications of eDNA data and can aid decision making by quantifying uncertainty in survey results.
Environmental DNA Sampling in Estuaries
Estuarine hydrology varies greatly and can be dynamic in space and time, creating challenges for eDNA detection and interpretation. In a large river-dominated estuary, physical processes such as variation in freshwater flow and tide are year-round drivers of chemical and biological processes (Kimmerer 2004). Smaller estuaries may see only seasonal freshwater input, resulting in high water residence times and periodic hypersaline conditions (Cloern et al. 2016a). Both unidirectional flow and tidal action in estuaries transport eDNA from its source, increasing the possibilities for eDNA dispersal from its origin. Particle tracking models have been used to characterize estuarine hydrology (e.g., https://water.ca.gov/Library/Modeling-and-Analysis/Bay-Delta-Region-models-and-tools/Delta-Simulation-Model-II), but studies integrating these models with eDNA have not yet been published in the context of estuaries. While water mixing and shorter water residence times may create the opportunity for essentially independent samples to be collected from the same location daily (Tillotson et al. 2018), drought conditions may alter this dynamic by increasing water residence times (Cloern et al. 2016b). Sampling design and interpretation of eDNA detection would greatly benefit from an understanding of estuarine hydrodynamics.
Estuaries experience high and variable turbidity more often than most freshwater and marine environments. Suspended particles may contain compounds that inhibit PCR (Matheson et al. 2010) and can limit water filtration volumes as filters become clogged with particulate matter; eDNA may also bind to particulate matter (Barnes et al. 2021). PCR inhibition can often be eliminated or significantly reduced using DNA extraction methods that include inhibition removal (e.g., DNeasy PowerWater or PowerSoil Pro Extraction (Qiagen, Valencia, CA USA)) or with a post-extraction inhibitor removal (e.g., OneStepPCR Inhibitor Removal Kit (Zymo Research, Irvine, CA, USA)). Filter clogging, on the other hand, remains a major challenge for eDNA sampling in turbid environments but can be ameliorated with specialized approaches such as multi-filter protocols (Hunter et al. 2019).
Although most published aquatic eDNA studies have been performed in freshwater and marine habitats, more estuarine-focused studies are starting to appear. For example, the eDNA (qPCR) detection probability of endangered tidewater goby (Eucyclogobius newberryi) was doubled compared with seine sampling in estuarine, lagoon, and slough habitats in coastal California and Oregon (Schmelzle and Kinziger 2016). In another study, three vulnerable manatee species (Trichechus spp.) were detected by eDNA qPCR and droplet digital PCR in multiple locations that included turbid estuary systems; eDNA detection estimates were found to be higher than aerial survey data (Hunter et al. 2018). Metabarcoding surveys of fish in the lower Hudson River Estuary, USA, had similar patterns of species detection (including seasonality and habitat preference) compared with 12 traditional monitoring surveys (Stoeckle et al. 2017). In a study that included three different river estuaries in the UK, the best-performing metabarcoding primer set/barcode (12S MiFish-U) detected more marine/estuarine fish species compared to traditional surveys at the estuarine sites; the authors noted that barcode and primer selection are critical for obtaining accurate and reproducible results (Collins et al. 2019). In the Pearl River Estuary in southern China, metabarcoding detected 32% more fish species compared with bottom trawling (Zou et al. 2020). Finally, a comprehensive metabarcoding analysis of 25 watersheds in Japan detected 290 fish species and reported that latitude and water temperature influence both river-mouth and coastal-sea fish community structure (Kume et al. 2021). Taken together, the results from these estuarine eDNA studies are highly encouraging and suggest that non-invasive eDNA methods can complement traditional survey methods. In support of this interpretation, a meta-analysis of published eDNA metabarcoding studies in natural aquatic environments found that, when analyzed collectively, eDNA metabarcoding and conventional surveys provided similar fish species richness estimates, including in estuaries (McElroy et al. 2020).
In addition to benchmarking eDNA methods to other survey methods, a handful of publications are starting to improve our understanding of some of the species-specific, environmental, and methodological considerations for eDNA in estuarine environments. For example, a study aiming to detect the invasive European green crab (Carcinus maenas) in a salt marsh system found that life stage is a critical determinant, with ovigerous individuals shedding significantly more eDNA than non-reproductive individuals (Crane et al. 2021). At the community level, salinity and temperature (used as a proxy for water mass) explained most of the variation in eDNA metabarcoding assemblages across sites and tides in the Hood Canal estuary near Seattle, USA (Kelly et al. 2018). Another study evaluated and optimized methods for Chinook salmon (Oncorhynchus tshawytscha) eDNA detection in the SFE using combined samples of estuarine water and water from tanks containing salmon (Sanches and Schreier 2020). In the large Elbe River estuary in Germany, eDNA metabarcoding revealed that tidal flows influenced species detection: downstream sampling locations were similar to sites further upstream during low tide, but species composition changed at the downstream sites at high tide (Schwentner et al. 2021). Other eDNA approaches such as indirect detection using sediment eDNA (e.g., Chariton et al. 2015; Lallias et al. 2015) and direct detection of plankton (Abad et al. 2016; Jungbluth et al. 2021) have been used in estuaries. Taken as a whole, the estuarine eDNA literature is limited, but growing. Additional work is needed, especially considering the wide diversity of estuary environments around the world, but the publications described above provide starting points for future studies.
Environmental DNA and Management Challenges in the San Francisco Estuary
Potential for eDNA Methods in the San Francisco Estuary
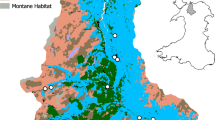
The San Francisco Estuary (SFE) in California, USA, is the largest estuary on the Pacific coast of North America, including a large inland delta (Fig. 1). Like many other estuaries located near urban centers, the SFE has been heavily altered by human activity. In this system, resource managers must balance the need for water exports to other parts of the state with the sustainment of the ecosystem for native endangered species (Luoma et al. 2015). Because of the many anthropomorphic alterations to the estuary and its connected watershed, there are specific challenges that need to be understood to improve its management. For example, the seasonal flow of freshwater into the estuary has become more variable and is severely reduced during periods of extreme drought. These fluctuations need to be managed with the extensive infrastructure consisting of pumps and canals in the south Delta that transport freshwater for agriculture in California’s Central Valley and drinking water for Southern California. Due in large part to water management in the Delta, a sharp decrease in abundance has been documented in several native and introduced fish species, termed the pelagic organism decline (Sommer et al. 2007), and some fishes, like the federally threatened delta smelt (Hypomesus transpacificus), are now perilously close to extinction (Hobbs et al. 2017). This loss is compounded by the multiple waves of invasive species that have entered the SFE (Simberloff 2021). To help address these challenges, a key SFE resource planning document, the 2017–2021 Science Action Agenda (https://scienceactionagenda.deltacouncil.ca.gov/) has prioritized modernization of monitoring as well as the development of tools for evaluating the success of habitat restoration programs.
The San Francisco Estuary (SFE) which includes a large inland delta. The figure shows water depth (blue shading), representative traditional (non-eDNA) monitoring locations (light gray dots), and wetland habitat restoration sites (green). There is extensive traditional monitoring throughout the region and eDNA monitoring could supplement these efforts, both concurrently to improve detection efficiency and also to expand monitoring both temporally and spatially in a cost- and resource-effective manner. Current traditional monitoring is more concentrated in the deepest areas of estuary, while restoration efforts are located in shallower regions. Environmental DNA (eDNA) surveys might be especially beneficial in the expansive shallow regions of the Delta. For shallow or marshy areas that cannot be sampled from shore, eDNA collection using small, shallow-draft boats like canoes or air boats would be ideal to avoid disturbing sediments and possibly resuspending settled eDNA
Environmental DNA detection methods could assist in modernization of monitoring in the SFE. Currently, several agencies at the state and federal level collectively operate multiple monitoring programs that provide essential data for SFE management, although some areas remain under-sampled (Stompe et al. 2020; Fig. 1). Species type and abundance is monitored by partially overlapping surveys using trawls, beach seines, gill nets, fyke traps, and screw traps. At this time, eDNA methods are likely to complement rather than replace all traditional monitoring, in part because some critical demographic information (e.g., organism size and developmental stage) cannot be determined by eDNA methods alone (Hansen et al. 2018). However, many species and habitats can be more effectively monitored using eDNA methods than with traditional methods, especially in habitats that are hard to access or may be missed due to sampling inefficiency. Furthermore, in addition to targeted (single species or a few taxa) eDNA assays, metabarcoding approaches could also complement existing monitoring and directives. This might include detection of fish, invertebrate, phytoplankton, and microbial assemblages by metabarcoding; assisting with planning and evaluation of large-scale management actions; and assessment of habitat restoration success. One of the benefits of aquatic eDNA sampling is the ability to sample for many different species that live in and around water simultaneously, potentially increasing sampling efficiency and use of limited agency resources. Another key benefit is that eDNA sampling is non-invasive, which can be especially relevant for listed species when take is a consideration.
With numerous and diverse taxa to survey, managers who work within the SFE often must prioritize resources based on requirements and mandates from government agencies. For example, recently, the California Department of Water Resources (DWR) received management mandates as part of their 2020 Incidental Take Permit (ITP) from the California Department of Fish and Wildlife to operate the State Water Project and deliver water to contractors across the state. According to this document, DWR must provide support, funding, and resources to long-term fish monitoring projects, including those focused on delta smelt, longfin smelt (Spirinchus thaleichthys), and Spring-run and Winter-run Chinook salmon (Oncorhynchus tshawytscha), as well as support reintroduction and restoration work. Other federal mandates that cover the endangered and threated SFE species listed above, as well as Southern Distinct Population Segment Green Sturgeon (Acipenser medirostris), include the 2019 U.S. Fish and Wildlife Service Biological Opinion, the 2019 U.S. Bureau of Reclamation Biological Assessment, and the 2019 National Marine Fisheries Service Biological Opinion. These designated species would be well-suited candidates for eDNA studies because of their designation across multiple conservation plans and monitoring directives.
Determining High-Priority SFE Species for eDNA Assessment
To identify monitoring needs for specific SFE species and taxonomic groups that could benefit from eDNA monitoring, we conducted an informal online survey of regional managers and scientists working in government agencies (Fig. 2 and Supplemental File). For the survey, we did not disclose that this was for assessing an application specific to eDNA detection of species because we did not want to bias responses from participants based on what they thought is possible to detect from eDNA. Respondents were asked to list up to 10 species in the SFE that they are most concerned with, in no particular order and of no particular origin, status, or taxa. We received responses from 37 individuals representing 10 different agencies, both state and federal (Supplemental File). A total of 77 different species were identified, with more specific details such as run type or distinct population segment added into the overall species count. The three most frequently suggested species were fishes: Chinook salmon (Oncorhynchus tshawytscha), delta smelt (Hypomesus transpacificus), and green sturgeon (Acipenser medirostris). Other entries were long-established invasive species in the SFE (e.g., Brazilian waterweed; Egeria densa) as well as newly arrived invaders like the semi-aquatic rodent Nutria (Myocastor coypus).
Environmental DNA (eDNA) assay readiness for top twenty species of interest in the San Francisco Estuary (SFE), as indicated from a survey of managers. Species (rectangles) are grouped by invasive and native and color-coded by broad taxonomic groupings. Line styles represent the readiness of the assay in the context of the SFE. Published assays (Baerwald et al. 2011; Smith et al. 2012; Brandl et al. 2015; Fujiwara et al. 2016; Anderson et al. 2018; Kurobe et al. 2018; Sanches and Schreier 2020) are indicated with symbols (legend in lower left). Note: the eDNA detection of green sturgeon (Acipenser medirostris) was in the Stanislaus River, a tributary of the San Joaquin River that flows into the SFE
Targeted eDNA assays have been developed and tested for a handful of key SFE species yet there remains untapped potential for assay development for many additional organisms. Of the 10 most frequently identified species of concern for managers, all except Nutria have a published qPCR assay, although most have not yet been tested in the context of eDNA detection. Some of these qPCR assays were originally developed for analysis of fish gut contents (Baerwald et al. 2011; Brandl et al. 2015) and might require further optimization for eDNA use. Additionally, CRISPR-based genetic detection assays for three morphologically similar osmerids co-occurring in the SFE, including delta smelt, have been created (Baerwald et al. 2020b). Environmental DNA qPCR assay optimization, in the context of the estuarine environment of the SFE, has been reported for Chinook salmon (Sanches and Schreier 2020). In addition, multiple publications have described in situ eDNA detections of several SFE species of concern, including green sturgeon in SFE tributaries by qPCR (Bergman et al. 2016; Anderson et al. 2018). Cyanobacteria, including toxic Microcystis spp. and other aquatic microorganisms, have also been detected in the SFE with qPCR assays validated for two Microcystis genotypes (Kurobe et al. 2018). Based on the responses from managers, we further identified the top 10 species of concern that do not yet have assays (Table 2). The list represents a broad cross section of taxa, including fish (N = 3), birds (N = 2), plants (N = 3), and mammals (N = 2). These species represent opportunities for researchers to develop and expand eDNA resources to more fully address SFE monitoring needs.
Additional Applications for eDNA Methods in the SFE
Environmental DNA monitoring is especially useful for detection of rare and cryptic species of concern because it is non-invasive and can be broadly applied in currently under-sampled environments or when species are difficult or intractable to survey (or even impossible if a take permit is not available) with traditional methods (Renan et al. 2017; Mauvisseau et al. 2019). For example, eDNA from the endangered semi-aquatic giant garter snake (Thamnophis gigas) was detected at sites in the SFE where trapping is logistically challenging and limited by low detection probability (Schumer et al. 2019b). From a habitat perspective, many marshes and wetlands adjoining San Pablo and Suisun Bays include extensive shallow regions that are inaccessible by boat and are problematic for some sampling gear (Fig. 1). Environmental DNA sampling, either from shore, with drones, or potentially from a shallow-draft boat, could expand monitoring capabilities in such areas. Some SFE wetlands are undergoing managed restoration to expand habitat and food resources for listed fishes, and regular biodiversity monitoring with eDNA metabarcoding in these areas could be implemented to help managers evaluate restoration success where few other monitoring methods are available.
Environmental DNA monitoring could also enable early detection of invasive species and track emerging animal diseases in the SFE. The Quagga mussel (Dreissena bugensis) and Zebra mussel (Dreissena polymorpha) are invasive mollusks found in some parts of California but have not yet been detected in the SFE (https://wildlife.ca.gov/Conservation/Invasives/Quagga-Mussels). Methods for qPCR-based eDNA detection of Zebra and Quagga mussels have been tested in other systems and are well validated (Sepulveda et al. 2020b). In freshwater waterways feeding into the SFE, qPCR-based eDNA detection has been demonstrated for multiple imperiled freshwater mussels (Anodonta spp., Gonidea angulata, and Margaritifera falcata) (Preece et al. 2020). One potential application for this or similar methods is detection of snake fungal disease caused by the fungus Ophidiomyces ophiodiicola, which has been increasing on the eastern USA (Lorch et al. 2016) and was detected in California in 2019 (California Department of Fish and Wildlife 2019).
Assemblage-level monitoring could be improved and expanded by using eDNA metabarcoding in the SFE. Current monitoring programs provide some information on fish assemblages, but there are inherent limitations from sampling gear that targets limited size classes and habitats, as well as general bias towards pelagic sampling in the SFE (Stompe et al. 2020). For more comprehensive SFE monitoring, it has been recommended that surveys should assess not only listed native fishes, but also full assemblages that include listed and unlisted native, as well as non-native, fishes (Dahm et al. 2019). Environmental DNA metabarcoding can potentially detect not only full fish assemblages, but also phytoplankton (including toxic Microcystis spp.), other bacteria of concern such as Enterococcus, zooplankton (Jungbluth et al. 2021), epibenthic crustaceans, bivalves, and aquatic vegetation such as the invasive water primrose Ludwigia. Some methodological testing and optimization (for example, determining primer sets that could efficiently detect organisms to the required taxonomic level) would be needed to apply this robustly for SFE management purposes, but there are published methods that provide a path forward for such “Tree of Life” ecosystem biomonitoring (Stat et al. 2017). For example, the California Environmental DNA (CALeDNA) project is large-scale, taxonomically broad biodiversity monitoring program that uses eDNA metabarcoding to detect microbes, fungi, plants, and animals across the state of California from soil and sediment samples (Meyer et al. 2021). Programs such as this, providing eDNA results as open data, can help show which substrates and molecular markers have been successfully used to detect target taxa and could inform how to efficiently implement eDNA metabarcoding biomonitoring in the SFE. Potential management questions that could benefit from eDNA-based sampling are detailed in Table 3.
General Recommendations for Conducting eDNA Studies in an Estuary
Based on the list of high-priority SFE species, studies on eDNA sampling protocol optimization can guide managers on how to begin sampling for these species and incorporating eDNA data into more routine monitoring. Researchers should work to quantify the impact of estuary conditions, like higher turbidity and variable salinity, on eDNA ecology and its detection efficiency. Species that prefer environments with higher turbidity, like the federally endangered delta smelt, may be more challenging to detect using eDNA due to particulate matter clogging filters (Egeter et al. 2018). Additional studies integrating hydrodynamic models, especially in tidal environments, will increase understanding of the impact of sampling location on detection probability and help determine whether near shore sampling is sufficient or if mid channel sampling from a boat is necessary. Finally, investment to build more local taxonomic knowledge and species-specific DNA reference libraries (e.g., https://research.nhm.org/disco/), especially for non-fish species of concern, will provide a foundation for new single-species assays and more accurate and comprehensive identification of sampled eDNA with metabarcoding.
Methods for eDNA detection that have been well validated in one habitat might require protocol adjustment for different habitats (e.g., estuaries) (Mize et al. 2019; Bedwell and Goldberg 2020). This is especially important for eDNA sampling in shallow wetlands, where it can be difficult to sample from marshy shores and the sampling can stir up sediment. Wetland-specific methodological adaptations, for example additional water pre-filtration steps or an adjustment in filter pore size, might be needed based on conditions like turbidity (Goldberg et al. 2018; Takasaki et al. 2021) or optimization of DNA purification protocols to release DNA bound to suspended particles (Lever et al. 2015). In addition, the unique environmental conditions in wetlands may alter eDNA dynamics and detections compared to other systems. For example, the low UV-B penetration below 50 cm in wetlands with high dissolved organic carbon (Arts et al. 2000) might slow UV-mediated eDNA degradation. In addition, both acidity (Seymour et al. 2018) and microbial activities (Shogren et al. 2018) accelerate eDNA degradation and are likely important factors for degradation of estuarine eDNA.
Recommendations for Enhancing Collaboration Between eDNA Scientists and Natural Resource Managers
Each step of eDNA collection, detection, and interpretation presents opportunities for increased communication, transparency, and collaboration between eDNA scientists and natural resource managers. Here, we put forward guidance based on a round table discussion between co-authors – which included representatives from both fields. Published templates for the types of collaborations we focus on here, describing approaches termed “translational ecology” (Enquist et al. 2017) and “adaptive management” (Gregory et al. 2006), among others, will help expand upon this guidance and provide additional context.
The first step for implementing eDNA methods for management purposes is to identify the research questions that eDNA data can address. Framing potential eDNA data in the context of existing methods and data is essential, and eDNA study design should consider the relative strengths and advantages of different methods and combine them cohesively with eDNA methods when possible. Table 3 provides potential research questions of general relevance to managers that eDNA data can answer. For example: where is an endangered species or species of management interest occurring? How has community composition changed after habitat restoration? Is an invasive species present in a specific watershed? Along with others listed in Table 3, these general research questions can also be expanded upon and customized for specific management needs. For example, for managers monitoring the specific location of the invasion front for an invasive species, and deciding if expensive eradication efforts are warranted, joint modeling of eDNA and traditional survey data can increase the precision of species density estimates at the leading edge of invasion (Keller et al. 2022). In some cases, combining eDNA and other methods (e.g., trapping) might be the optimal approach; in others, eDNA data could be useful as an initial step to determine if traditional monitoring is needed (Table 3). Alternatively, when eDNA methods have significant logistical advantages or are much less costly than traditional monitoring, an eDNA-alone approach might be justified.
After identification of the research question and the relevance of eDNA methods for answering it, we recommend that eDNA scientists include managers in all steps of planning and implementing eDNA methods. These steps include study conceptualization, determination of what constitutes a positive detection, and what actions will be taken for positive or negative detections, field work, laboratory work, data analysis, and interpretation (Fig. 3). To promote communication, we recommend holding regular meetings between eDNA scientists and managers when feasible. To facilitate these meetings and enhance communication in general, one person could be designated as a liaison who would be accountable for maintaining consistent communication through updates and meetings. A major goal of these meetings is to build trust; it is important to set realistic expectations about what data eDNA methods can provide. This communication can also be very important when the stakes of the outcome are high. In such cases, creating a clear and coordinated plan for how and when results are communicated to the media and public can also increase trust in the process (e.g., Abbott et al. 2021). Such methods are employed in the use of eDNA detections of Asian carp in the North American Great Lakes (Jerde 2019). Environmental DNA assay validation scales, such as those suggested by Thalinger et al. (2021), provide one objective framework to judge assay readiness. Additionally, for commonly used single-species assays, cross-laboratory comparisons could help with understanding repeatability, reproducibility, and accuracy of quantification (Sepulveda et al. 2020b). For metabarcoding, bioinformatics methods have the potential to be a “black box” where the details of data analysis pipelines are less accessible to non-specialists, while the bioinformatics personnel may not have the expertise to determine when an apparent detection is questionable and requires additional review. Collaborations and discussions about interpretation of taxonomic assignments for DNA sequences from metabarcoding data would therefore assist in building consensus. More generally, the creation of a regional eDNA science advisory panel (for example, for the SFE), composed of outside experts, could advance the application of eDNA methods by acting as a sounding board for ideas and providing recommendations. Finally, all eDNA detection studies should be conducted using an open data framework to ensure transparency and reproducibility (Baerwald et al. 2020a; Goldberg et al. 2016).
Environmental DNA (eDNA) workflow and opportunities for enhancing communication between eDNA scientists and managers. The top shows two major cycles in the workflow: a development and a deployment cycle. The text at the bottom provides recommendations for each step in the workflow to increase communication and build confidence in the application of eDNA methods to meet management needs
Conclusions
How can eDNA data be used for management decisions in estuaries? There is now a consensus that eDNA methods are reliable to determine presence/non-detection and are especially useful for rare and invasive species that are hard to detect with traditional methods (Pfleger et al. 2016). Because eDNA methods involve indirect detection, there are some limitations that are important to keep in mind. Some of the most important considerations are (1) eDNA does not give direct information about organism size or absolute abundance; (2) detections have some spatial and temporal ambiguity; (3) demographic data such as age and life stage cannot be determined at this time; and (4) assays developed in one natural system may not work in others because of differences in biodiversity, chemical inhibitors, and hydrogeochemical differences, and thus require pilot testing. For metabarcoding studies, ambiguous or incomplete sequence data can limit species-level identification. With these caveats in mind, it is notable that well-validated eDNA methods are reliable and meet the standards for scientific evidence in some US courts (Sepulveda et al. 2020c). For some applications, such as estimating species richness or delimiting the spatial extent of a species or community, eDNA may outperform conventional approaches in a cost effective way (Evans et al. 2017). When possible, eDNA data should be benchmarked to traditional methods, but they are fundamentally different methods with divergent pros and cons and should not be expected to mirror one another exactly, just as different traditional surveys and gears are not perfectly congruent (Shelton et al. 2019).
Several reviews have addressed application of eDNA methods specifically to fisheries management (Hansen et al. 2018; Jerde 2019). Like all sampling methods, eDNA is subject to potential errors. Imprecision in spatiotemporal inference, for example detecting eDNA that has been transported away from its origin, is one source of error. Models that incorporate data on eDNA particle state with hydrology provide a potential solution (Pont et al. 2018; Carraro et al. 2020), although more work is needed in estuaries. Additional species-specific information on shedding rates based on life cycle and habitat-specific studies of eDNA particle state, transport, and degradation will lay the foundation for greater integration of eDNA data into management decisions. Experiments to determine the relationship between eDNA concentration, biomass, and abundance should also be prioritized for species where this information is needed for management. For invasive species, there might be additional considerations for reporting data and for how managers can interpret putative positive eDNA detections (Jerde 2019; Sepulveda et al. 2020c). Since a positive detection might have wide-reaching consequences, one option is to use a Management (or Decision) Support Tree (Sepulveda et al. 2020c; Welsh et al. 2020; Abbott et al. 2021). This approach (1) evaluates the likelihood that a target organism could be present in the system based on local environmental information; (2) evaluates the strength of the eDNA evidence; (3) assigns a detection level (e.g., 1 weakest–4 strongest) to positive detections; and (4) recommends specific actions based on detection level.
Finally, eDNA data are already being incorporated into management decision making. For example, the European Union is currently using eDNA for indicator and invasive species (Andersen et al. 2016). In the US Midwest, the US Fish and Wildlife Service leads an eDNA surveillance program to detect invasive Asian carp that has collected over 30,000 water samples (https://www.fws.gov/midwest/Fisheries/eDNA.html). Their program has created a standard operating protocol (SOP) for disseminating eDNA data (https://www.fws.gov/midwest/Fisheries/eDNA/documents/SOP-eDNA-Results.pdf) that provides a model for other agencies considering eDNA for early detection of invasive species. With advancements and standardization in methodology, broad understanding of strengths and limitations, and enhanced communication, eDNA monitoring and biodiversity data will increasingly support management decisions in estuaries and beyond.
References
Abad, D., A. Albaina, M. Aguirre, et al. 2016. Is metabarcoding suitable for estuarine plankton monitoring? A comparative study with microscopy. Marine Biology 163: 1–13. https://doi.org/10.1007/s00227-016-2920-0.
Abbott, C., M. Coulson, N. Gagné, et al. 2021. Guidance on the use of targeted environmental DNA (eDNA) analysis for the management of aquatic invasive species and species at risk. Canadian Science Advisory Secretariat, 2021/019(March), 46.
Andersen, J., E. Kallenbach, M. Hesselsoe, and S.W. Knudsen. 2016. Steps toward nation-wide monitoring of non-indigenous species in Danish marine waters under the Marine Strategy Framework Directive. NIVA-rapport. 122 p. Norsk institutt for vannforskning.
Anderson, J.T., G. Schumer, P.J. Anders, et al. 2018. Confirmed observation: A north American green sturgeon acipenser medirostris recorded in the Stanislaus river, California. Journal of Fish and Wildlife Management 9: 624–630. https://doi.org/10.3996/012018-JFWM-006.
Andruszkiewicz, E., W.G. Zhang, A. Lavery, and A. Govindarajan. 2020. Environmental DNA shedding and decay rates from diverse animal forms and thermal regimes. Environmental DNA 3: 492–514. https://doi.org/10.1002/edn3.141.
Andruszkiewicz, E.A., J.R. Koseff, O.B. Fringer, et al. 2019. Modeling environmental DNA transport in the coastal ocean using Lagrangian particle tracking. Frontiers in Marine Science 6: 1–14. https://doi.org/10.3389/fmars.2019.00477.
Arts, M.T., R.D. Robarts, F. Kasai, et al. 2000. The attenuation of ultraviolet radiation in high dissolved organic carbon waters of wetlands and lakes on the northern Great Plains. Limnology and Oceanography 45: 292–299. https://doi.org/10.4319/lo.2000.45.2.0292.
Baerwald, M., B. Davis, S. Lesmeister, et al. 2020a. An open data framework for the San Francisco estuary. San Francisco Estuary and Watershed Science 18: 2. https://doi.org/10.15447/sfews.2020v18iss2art1.
Baerwald, M.R., A.M. Goodbla, R.P. Nagarajan, et al. 2020b. Rapid and accurate species identification for ecological studies and monitoring using CRISPR-based SHERLOCK. Molecular Ecology Resources 20: 961–970. https://doi.org/10.1111/1755-0998.13186.
Baerwald, M.R., G. Schumer, B.M. Schreier, and B. May. 2011. TaqMan assays for the genetic identification of delta smelt (Hypomesus transpacificus) and Wakasagi smelt (Hypomesus nipponensis). Molecular Ecology Resources 11: 784–785. https://doi.org/10.1111/j.1755-0998.2011.03011.x.
Baker, C.S., D. Steel, S. Nieukirk, and H. Klinck. 2018. Environmental DNA (eDNA) from the wake of the whales: Droplet digital PCR for detection and species identification. Frontiers in Marine Science 5: 1–11. https://doi.org/10.3389/fmars.2018.00133.
Barnes, M.A., W.L. Chadderton, C.L. Jerde, et al. 2021. Environmental conditions influence eDNA particle size distribution in aquatic systems. Environmental DNA 3: 643–653. https://doi.org/10.1002/edn3.160.
Barnes, M.A., and C.R. Turner. 2016. The ecology of environmental DNA and implications for conservation genetics. Conservation Genetics 17: 1–17. https://doi.org/10.1007/s10592-015-0775-4.
Barnes, M.A., C.R. Turner, C.L. Jerde, et al. 2014. Environmental conditions influence eDNA persistence in aquatic systems. Environmental Science and Technology 48: 1819–1827. https://doi.org/10.1021/es404734p.
Bedwell, M.E., and C.S. Goldberg. 2020. Spatial and temporal patterns of environmental DNA detection to inform sampling protocols in lentic and lotic systems. Ecology and Evolution 10: 1602–1612. https://doi.org/10.1002/ece3.6014.
Bergman, P.S., G. Schumer, S. Blankenship, and E. Campbell. 2016. Detection of adult green sturgeon using environmental DNA analysis. PLoS One 11: 1–8. https://doi.org/10.1371/journal.pone.0153500.
Brandl, S., G. Schumer, B.M. Schreier, et al. 2015. Ten real-time PCR assays for detection of fish predation at the community level in the San Francisco Estuary-Delta. Molecular Ecology Resources 15: 278–284. https://doi.org/10.1111/1755-0998.12305.
Bustin, S.A., V. Benes, J.A. Garson, et al. 2009. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55: 611–622. https://doi.org/10.1373/clinchem.2008.112797.
California Department of Fish and Wildlife. 2019. Snake fungal disease detected in California. https://cdfgnews.wordpress.com/2019/11/05/snake-fungal-disease-detected-in-california/. Accessed 11 Jun 2020.
Carraro, L., E. Mächler, R. Wüthrich, and F. Altermatt. 2020. Environmental DNA allows upscaling spatial patterns of biodiversity in freshwater ecosystems. Nature Communications 11: 1–12. https://doi.org/10.1038/s41467-020-17337-8.
Chariton, A.A., S. Stephenson, M.J. Morgan, et al. 2015. Metabarcoding of benthic eukaryote communities predicts the ecological condition of estuaries. Environmental Pollution 203: 165–174. https://doi.org/10.1016/j.envpol.2015.03.047.
Cloern, J.E., P.C. Abreu, J. Carstensen, et al. 2016a. Human activities and climate variability drive fast-paced change across the world’s estuarine-coastal ecosystems. Global Change Biology 22: 513–529. https://doi.org/10.1111/gcb.13059.
Cloern, J.E., P.J. Barnard, E. Beller, et al. 2016b. Estuaries: Life on the edge. In Ecosystems of California, ed. H. Mooney and E. Zavaleta, 133–137. University of California Press.
Collins, R.A., J. Bakker, O.S. Wangensteen, et al. 2019. Non-specific amplification compromises environmental DNA metabarcoding with COI. Methods in Ecology and Evolution. https://doi.org/10.1111/2041-210x.13276.
Connon, R.E., L.S. D’Abronzo, N.J. Hostetter, et al. 2012. Transcription profiling in environmental diagnostics: Health assessments in Columbia River basin steelhead (Oncorhynchus mykiss). Environmental Science and Technology 46: 6081–6087. https://doi.org/10.1021/es3005128.
Crane, L.C., J.S. Goldstein, D.W. Thomas, et al. 2021. Effects of life stage on eDNA detection of the invasive European green crab (Carcinus maenas) in estuarine systems. Ecological Indicators 124. https://doi.org/10.1016/j.ecolind.2021.107412.
Cristescu, M.E. 2019. Can Environmental RNA revolutionize biodiversity science? Trends in Ecology & Evolution 34: 694–697. https://doi.org/10.1016/j.tree.2019.05.003.
Dahm, C., W. Kimmerer, J. Korman, et al. 2019. Developing biological goals for the Bay-Delta Plan : Concepts and ideas from an independent scientific advisory panel. A final report to the Delta Science Program.
Darling, J.A. C.L. Jerde, and A.J. Sepulveda. 2021. What do you mean by false positive? Environmental DNA 3: 877–1046. https://doi.org/10.1002/edn3.194.
Darling, J.A., and A.R. Mahon. 2011. From molecules to management: Adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environmental Research 111: 978–988. https://doi.org/10.1016/j.envres.2011.02.001.
Deiner, K., and F. Altermatt. 2014. Transport distance of invertebrate environmental DNA in a natural river. PLoS One 9 (2). https://doi.org/10.1371/journal.pone.0088786
Deiner, K., H. Yamanaka, and L. Bernatchez. 2021. The future of biodiversity monitoring and conservation utilizing environmental DNA. Environmental DNA 3: 3–7. https://doi.org/10.1002/edn3.178.
Doi, H., T. Takahara, T. Minamoto, et al. 2015. Droplet digital polymerase chain reaction (PCR) outperforms real-time PCR in the detection of environmental DNA from an invasive fish species. Environmental Science and Technology 49: 5601–5608. https://doi.org/10.1021/acs.est.5b00253.
Dorazio, R.M., and R.A. Erickson. 2018. ednaoccupancy: An r package for multiscale occupancy modelling of environmental DNA data. Molecular Ecology Resources 18: 368–380. https://doi.org/10.1111/1755-0998.12735.
Dunker, K.J., A.J. Sepulveda, R.L. Massengill, et al. 2016. Potential of environmental DNA to evaluate northern pike (Esox lucius) eradication efforts: An experimental test and case study. PLoS One 11: 1–21. https://doi.org/10.1371/journal.pone.0162277.
Eble, J.A., T.S. Daly-Engel, J.D. DiBattista, et al. 2020. Marine environmental DNA: Approaches, applications, and opportunities, 1st edn. Elsevier Ltd.
Egeter, B., S. Peixoto, J.C. Brito, et al. 2018. Challenges for assessing vertebrate diversity in turbid Saharan water-bodies using environmental DNA. Genome 61: 807–814. https://doi.org/10.1139/gen-2018-0071.
Eichmiller, J.J., S.E. Best, and P.W. Sorensen. 2016. Effects of temperature and trophic state on degradation of environmental DNA in lake water. Environmental Science and Technology 50: 1859–1867. https://doi.org/10.1021/acs.est.5b05672.
Enquist, C.A.F., S.T. Jackson, G.M. Garfin, et al. 2017. Foundations of translational ecology. Frontiers in Ecology and the Environment 15: 541–550. https://doi.org/10.1002/fee.1733.
Espe, M.B., M. Johnston, S.M. Blankenship, et al. 2022. The artemis package for environmental DNA analysis in R. Environmental DNA. https://doi.org/10.1002/edn3.277.
Evans, N.T., P.D. Shirey, J.G. Wieringa, et al. 2017. Comparative cost and effort of fish distribution detection via environmental DNA analysis and electrofishing. Fisheries 42: 90–99. https://doi.org/10.1080/03632415.2017.1276329.
Ficetola, G.F., C. Miaud, F. Pompanon, and P. Taberlet. 2008. Species detection using environmental DNA from water samples. Biology Letters 4: 423–425. https://doi.org/10.1098/rsbl.2008.0118.
Forootan, A., R. Sjöback, J. Björkman, et al. 2017. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR). Biomolecular Detection and Quantification 12: 1–6. https://doi.org/10.1016/j.bdq.2017.04.001.
Fremier, A.K., K.M. Strickler, J. Parzych, et al. 2019. Stream transport and retention of environmental DNA pulse releases in relation to hydrogeomorphic scaling factors. Environmental Science and Technology 53: 6640–6649. https://doi.org/10.1021/acs.est.8b06829.
Fujiwara, A., S. Matsuhashi, H. Doi, et al. 2016. Use of environmental DNA to survey the distribution of an invasive submerged plant in ponds. Freshwater Science 35: 748–754. https://doi.org/10.1086/685882.
Goldberg, C.S., K.M. Strickler, and A.K. Fremier. 2018. Degradation and dispersion limit environmental DNA detection of rare amphibians in wetlands: Increasing efficacy of sampling designs. Science of the Total Environment 633: 695–703. https://doi.org/10.1016/j.scitotenv.2018.02.295.
Goldberg, C.S., C.R. Turner, K. Deiner, et al. 2016. Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods in Ecology and Evolution 7: 1299–1307. https://doi.org/10.1111/2041-210X.12595.
Gregory, R., D. Ohlson, and J. Arvai. 2006. Deconstructing adaptive management: Criteria for applications to environmental management. Ecological Applications 16: 2411–2425. https://doi.org/10.1890/1051-0761(2006)016[2411:DAMCFA]2.0.CO;2.
Hansen, B.K., D. Bekkevold, L.W. Clausen, and E.E. Nielsen. 2018. The sceptical optimist: Challenges and perspectives for the application of environmental DNA in marine fisheries. Fish and Fisheries 19: 751–768. https://doi.org/10.1111/faf.12286.
Harrison, J.B., J.M. Sunday, and S.M. Rogers. 2019. Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proceeding of the Royal Society B Biological Science 286. https://doi.org/10.1098/rspb.2019.1409.
Hobbs, J., P. Moyle, N. Fangue, and R.E. Connon. 2017. Is extinction inevitable for delta smelt and longfin smelt? An opinion and recommendations for recovery. San Francisco Estuary and Watershed Science 15: 2. https://doi.org/10.15447/sfews.2017v15iss2art2.
Hunter, M.E., R.M. Dorazio, J.S.S. Butterfield, et al. 2017. Detection limits of quantitative and digital PCR assays and their influence in presence–absence surveys of environmental DNA. Molecular Ecology Resources 17: 221–229. https://doi.org/10.1111/1755-0998.12619.
Hunter, M.E., J.A. Ferrante, G. Meigs-Friend, and A. Ulmer. 2019. Improving eDNA yield and inhibitor reduction through increased water volumes and multi-filter isolation techniques. Scientific Reports 9(1). https://doi.org/10.1038/s41598-019-40977-w.
Hunter, M.E., G. Meigs-Friend, J.A. Ferrante, et al. 2018. Surveys of environmental DNA (eDNA): A new approach to estimate occurrence in vulnerable manatee populations. Endangered Species Research 35: 101–111. https://doi.org/10.3354/esr00880.
Hunter, M.E., S.J. Oyler-McCance, R.M. Dorazio, et al. 2015. Environmental DNA (eDNA) sampling improves occurrence and detection estimates of invasive Burmese pythons. PLoS One 10: 1–17. https://doi.org/10.1371/journal.pone.0121655.
Jane, S.F., T.M. Wilcox, K.S. Mckelvey, et al. 2015. Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Molecular Ecology Resources 15: 216–227. https://doi.org/10.1111/1755-0998.12285.
Jerde, C.L. 2019. Can we manage fisheries with the inherent uncertainty from eDNA? Journal of Fish Biology 98: 341–353. https://doi.org/10.1111/jfb.14218.
Jerde, C.L., B.P. Olds, A.J. Shogren, et al. 2016. Influence of stream bottom substrate on retention and transport of vertebrate environmental DNA. Environmental Science and Technology 50: 8770–8779. https://doi.org/10.1021/acs.est.6b01761.
Jo, T., H. Murakami, S. Yamamoto, et al. 2019. Effect of water temperature and fish biomass on environmental DNA shedding, degradation, and size distribution. Ecology and Evolution 9: 1135–1146. https://doi.org/10.1002/ece3.4802.
Jungbluth, M.J., J. Burns, L. Grimaldo, et al. 2021. Feeding habits and novel prey of larval fishes in the northern San Francisco Estuary. Environmental DNA 3(6): 1059–1080. https://doi.org/10.1002/edn3.226.
Keller, A.G., E.W. Grason, P.S. McDonald, et al. 2022. Tracking an invasion front with environmental DNA. Ecological Applications e2561. https://doi.org/10.1002/eap.2561.
Kelly, R.P., R. Gallego, and E. Jacobs-Palme. 2018. The effect of tides on nearshore environmental DNA. Peer Journal (3). https://doi.org/10.7717/peerj.4521.
Kelly, R.P., A.O. Shelton, and R. Gallego. 2019. Understanding PCR processes to draw meaningful conclusions from environmental DNA studies. Scientific Reports 9: 1–14. https://doi.org/10.1038/s41598-019-48546-x.
Kimmerer, W.J. 2004. Open Water processes of the San Francisco estuary: From physical forcing to biological responses. San Francisco Estuary and Watershed Science 2(1). https://doi.org/10.15447/sfews.2004v2iss1art1.
Klymus, K.E., C.M. Merkes, M.J. Allison, et al. 2020. Reporting the limits of detection and quantification for environmental DNA assays. Environmental DNA 2(3): 271–282. https://doi.org/10.1002/edn3.29.
Klymus, K.E., C.A. Richter, D.C. Chapman, and C. Paukert. 2015. Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biological Conservation 183: 77–84. https://doi.org/10.1016/j.biocon.2014.11.020.
Kumar, G., J.E. Eble, and M.R. Gaither. 2020. A practical guide to sample preservation and pre-PCR processing of aquatic environmental DNA. Molecular Ecology Resources 20: 29–39. https://doi.org/10.1111/1755-0998.13107.
Kume, M., E. Lavergne, H. Ahn, et al. 2021. Factors structuring estuarine and coastal fish communities across Japan using environmental DNA metabarcoding. Ecological Indicators 121: 107216. https://doi.org/10.1016/j.ecolind.2020.107216.
Kurobe, T., P.W. Lehman, B.G. Hammock, et al. 2018. Biodiversity of cyanobacteria and other aquatic microorganisms across a freshwater to brackish water gradient determined by shotgun metagenomic sequencing analysis in the San Francisco Estuary, USA. PLoS One 13(9). https://doi.org/10.1371/journal.pone.0203953.
Lacoursière-Roussel, A., M. Rosabal, and L. Bernatchez. 2016. Estimating fish abundance and biomass from eDNA concentrations: Variability among capture methods and environmental conditions. Molecular Ecology Resources 16: 1401–1414. https://doi.org/10.1111/1755-0998.12522.
Lacoursière-Roussel, A., and K. Deiner. 2019. Environmental DNA is not the tool by itself. Journal of Fish Biology 98: 383–386. https://doi.org/10.1111/jfb.14177.
Lallias, D., J.G. Hiddink, V.G. Fonseca, et al. 2015. Environmental metabarcoding reveals heterogeneous drivers of microbial eukaryote diversity in contrasting estuarine ecosystems. ISME Journal 9: 1208–1221. https://doi.org/10.1038/ismej.2014.213.
Laporte, M., B. Bougas, G. Côté, et al. 2020. Caged fish experiment and hydrodynamic bidimensional modeling highlight the importance to consider 2D dispersion in fluvial environmental DNA studies. Environmental DNA 3(2): 362–372. https://doi.org/10.1002/edn3.88.
Leray, M., J.Y. Yang, C.P. Meyer, et al. 2013. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Frontiers in Zoology 10: 1–14. https://doi.org/10.1186/1742-9994-10-34.
Lesperance, M.L., M.J. Allison, L.C. Bergman, et al. 2021. A statistical model for calibration and computation of detection and quantification limits for low copy number environmental DNA samples. Environmental DNA 3(5): 970–981. https://doi.org/10.1002/edn3.220.
Lever, M.A., A. Torti, P. Eickenbusch, et al. 2015. A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types. Frontiers in Microbiology 6(MAY). https://doi.org/10.3389/fmicb.2015.00476.
Lin, M., A.L. Simons, R.J. Harrigan, et al. 2021. Landscape analyses using eDNA metabarcoding and Earth observation predict community biodiversity in California. Ecological Applications 31(6). https://doi.org/10.1002/eap.2379.
Lorch, J.M., S. Knowles, J.S. Lankton, et al. 2016. Snake fungal disease: An emerging threat to wild snakes. Philosophical Transaction of the Royal Society B Biological Sciences 371. https://doi.org/10.1098/rstb.2015.0457.
Luoma, S.N., C.N. Dahm, M. Healey, and J.N. Moore. 2015. Challenges facing the Sacramento-San Joaquin Delta: Complex, chaotic, or simply cantankerous? San Francisco Estuary and Watershed Science 13. https://doi.org/10.15447/sfews.2015v13iss3art7.
Mahon, A.R., C.L. Jerde, M. Galaska, et al. 2013. Validation of eDNA surveillance sensitivity for detection of Asian carps in controlled and field experiments. PLoS One 8(3). https://doi.org/10.1371/journal.pone.0058316.
Matheson, C.D., C. Gurney, N. Esau, and R. Lehto. 2010. Assessing PCR inhibition from humic substances. Open Enzyme Inhibition Journal 3: 38–45. https://doi.org/10.2174/1874940201003010038.
Mauvisseau, Q., J. Davy-Bowker, M. Bulling, et al. 2019. Combining ddPCR and environmental DNA to improve detection capabilities of a critically endangered freshwater invertebrate. Scientific Reports 9: 1–9. https://doi.org/10.1038/s41598-019-50571-9.
McElroy, M.E., T.L. Dressler, G.C. Titcomb, et al. 2020. Calibrating Environmental DNA Metabarcoding to Conventional Surveys for Measuring Fish Species Richness. Frontiers in Ecology and Evolution 8. https://doi.org/10.3389/fevo.2020.00276.
McLaren, M.R., A.D. Willis, and B.J. Callahan. 2019. Consistent and correctable bias in metagenomic sequencing experiments. eLife 8: 1–37. https://doi.org/10.7554/elife.46923.
Meyer, R.S., M.M. Ramos, M. Lin, et al. 2021. The CALeDNA program: Citizen scientists and researchers inventory California’s biodiversity. California Agriculture 75: 20–32. https://doi.org/10.3733/ca.2021a0001.
Miya, M., Y. Sato, T. Fukunaga, et al. 2015. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. Royal Society Open Science 2(7). https://doi.org/10.1098/rsos.150088.
Mize, E.L., R.A. Erickson, C.M. Merkes, et al. 2019. Refinement of eDNA as an early monitoring tool at the landscape-level: Study design considerations. Ecological Applications 29: 1374–1388. https://doi.org/10.1002/eap.1951.
Mordecai, G.J., K.M. Miller, E. Di Cicco, et al. 2019. Endangered wild salmon infected by newly discovered viruses. eLife 8. https://doi.org/10.7554/eLife.47615.
Moushomi, R., G. Wilgar, G. Carvalho, et al. 2019. Environmental DNA size sorting and degradation experiment indicates the state of Daphnia magna mitochondrial and nuclear eDNA is subcellular. Scientific Reports 9(1). https://doi.org/10.1038/s41598-019-48984-7.
Pawlowski, J., L. Apothéloz-Perret-Gentil, and F. Altermatt. 2020. Environmental DNA: What’s behind the term? Clarifying the terminology and recommendations for its future use in biomonitoring. Molecular Ecology 29: 4258–4264. https://doi.org/10.1111/mec.15643.
Pawlowski, J., L. Apothéloz-Perret-Gentil, and F. Altermatt. 2021. Environmental versus extra-organismal DNA. Molecular Ecology 30: 4606–4607. https://doi.org/10.1111/mec.16144.
Pfleger, M.O., S.J. Rider, C.E. Johnston, and A.M. Janosik. 2016. Saving the doomed: Using eDNA to aid in detection of rare sturgeon for conservation (Acipenseridae). Global Ecology and Conservation 8: 99–107. https://doi.org/10.1016/j.gecco.2016.08.008.
Pilliod, D.S., C.S. Goldberg, R.S. Arkle, and L.P. Waits. 2014. Factors influencing detection of eDNA from a stream-dwelling amphibian. Molecular Ecology Resources 14: 109–116. https://doi.org/10.1111/1755-0998.12159.
Pilliod, D.S., C.S. Goldberg, M.B. Laramie, and L.P. Waits. 2013. Application of environmental DNA for inventory and monitoring of aquatic species. U.S. Geological Survey, Fact Sheet 2012-3146. https://doi.org/10.3133/fs20123146.
Pochon, X., A. Zaiko, L.M. Fletcher, et al. 2017. Wanted dead or alive? Using metabarcoding of environmental DNA and RNA to distinguish living assemblages for biosecurity applications. PLoS One 12(11). https://doi.org/10.1371/journal.pone.0187636.
Pont, D., M. Rocle, A. Valentini, et al. 2018. Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Scientific Reports 8: 1–13. https://doi.org/10.1038/s41598-018-28424-8.
Port, J.A., J.L. O’Donnell, O.C. Romero-Maraccini, et al. 2016. Assessing vertebrate biodiversity in a kelp forest ecosystem using environmental DNA. Molecular Ecology 25: 527–541. https://doi.org/10.1111/mec.13481.
Preece, E.P., M. Bryan, S.M. Mapes, et al. 2020. Monitoring for freshwater mussel presence in rivers using environmental DNA. Environmental DNA 3(3): 591–604. https://doi.org/10.1002/edn3.156.
Rees, H.C., B.C. Maddison, D.J. Middleditch, et al. 2014. The detection of aquatic animal species using environmental DNA - A review of eDNA as a survey tool in ecology. Journal of Applied Ecology 51: 1450–1459. https://doi.org/10.1111/1365-2664.12306.
Renan, S., S. Gafny, R.G.B. Perl, et al. 2017. Living quarters of a living fossil – Uncovering the current distribution pattern of the rediscovered Hula painted frog (Latonia nigriventer) using environmental DNA. Molecular Ecology 26: 6801–6812. https://doi.org/10.1111/mec.14420.
Rodriguez-Ezpeleta, N., O. Morissette, C.W. Bean, et al. 2021. Trade-offs between reducing complex terminology and producing accurate interpretations from environmental DNA: Comment on “Environmental DNA: What’s behind the term?” by Pawlowski et al. 2020. Molecular Ecology. https://doi.org/10.1111/mec.15942.
Rourke, M.L., A.M. Fowler, J.M. Hughes, et al. 2022. Environmental DNA (eDNA) as a tool for assessing fish biomass : A review of approaches and future considerations for resource surveys. Environmental DNA 4: 9–33. https://doi.org/10.1002/edn3.185.
Royle, J.A., J.D. Nichols, M. Kéry, E. and Ranta. 2005. Modelling occurrence and abundance of species when detection is imperfect. Oikos 100(2): 353–359. https://doi.org/10.1111/j.0030-1299.2005.13534.x.
Sanches, T.M., and A.D. Schreier. 2020. Optimizing an eDNA protocol for estuarine environments: Balancing sensitivity, cost and time. PLoS One 15: 1–18. https://doi.org/10.1371/journal.pone.0233522.
Sassoubre, L.M., K.M. Yamahara, L.D. Gardner, et al. 2016. Quantification of environmental DNA (eDNA) shedding and decay rates for three marine fish. Environmental Science and Technology 50: 10456–10464. https://doi.org/10.1021/acs.est.6b03114.
Schenekar, T., M. Schletterer, L.A. Lecaudey, and S.J. Weiss. 2020. Reference databases, primer choice, and assay sensitivity for environmental metabarcoding: Lessons learnt from a re-evaluation of an eDNA fish assessment in the Volga headwaters. River Research and Applications 36: 1004–1013. https://doi.org/10.1002/rra.3610.
Schill, W.B. 2020. Capture of environmental dna (Edna) from water samples by flocculation. Journal of Visualized Experiments 2020: 1–5. https://doi.org/10.3791/60967.
Schmelzle, M.C., and A.P. Kinziger. 2016. Using occupancy modelling to compare environmental DNA to traditional field methods for regional-scale monitoring of an endangered aquatic species. Molecular Ecology Resources 16: 895–908. https://doi.org/10.1111/1755-0998.12501.
Schmidt, B.R., M. Kéry, S. Ursenbacher, et al. 2013. Site occupancy models in the analysis of environmental DNA presence/absence surveys: A case study of an emerging amphibian pathogen. Methods in Ecology and Evolution 4: 646–653. https://doi.org/10.1111/2041-210X.12052.
Schumer, G., K. Crowley, E. Maltz, et al. 2019a. Utilizing environmental DNA for fish eradication effectiveness monitoring in streams. Biological Invasions 21: 3415–3426. https://doi.org/10.1007/s10530-019-02056-z.
Schumer, G., E.C. Hansen, P.J. Anders, and S.M. Blankenship. 2019b. Development of a quantitative polymerase chain reaction assay and environmental DNA sampling methods for Giant Gartersnake (Thamnophis gigas). PLoS One 14: 1–13. https://doi.org/10.1371/journal.pone.0222493.
Schwentner, M., R. Zahiri, S. Yamamoto, et al. 2021. eDNA as a tool for non-invasive monitoring of the fauna of a turbid, well-mixed system, the Elbe estuary in Germany. PLoS One 16: 1–16. https://doi.org/10.1371/journal.pone.0250452.
Sepulveda, A.J., R. Al-Chokhachy, M.B. Laramie, et al. 2021. It’s complicated. Environmental dna as a predictor of trout and char abundance in streams. Canadian Journal of Fisheries and Aquatic Sciences 78: 422–432. https://doi.org/10.1139/cjfas-2020-0182.
Sepulveda, A.J., P.R. Hutchins, M. Forstchen, et al. 2020a. The elephant in the lab (and field): Contamination in aquatic environmental DNA studies. Frontiers in Ecology and Evolution 8: 1–12. https://doi.org/10.3389/fevo.2020.609973.
Sepulveda, A.J., P.R. Hutchins, C. Jackson, et al. 2020b. A round-robin evaluation of the repeatability and reproducibility of environmental DNA assays for dreissenid mussels. Environmental DNA 2(4): 446–459. https://doi.org/10.1002/edn3.68.
Sepulveda, A.J., P.R. Hutchins, R.L. Massengill, and K.J. Dunker. 2018. Tradeoffs of a portable, field-based environmental DNA platform for detecting invasive northern pike (Esox lucius) in Alaska. Management of Biological Invasions 9: 253–258. https://doi.org/10.3391/mbi.2018.9.3.07.
Sepulveda, A.J., N.M. Nelson, C.L. Jerde, and G. Luikart. 2020c. Are environmental DNA methods ready for aquatic invasive species management? Trends in Ecology & Evolution 35: 668–678. https://doi.org/10.1016/j.tree.2020.03.011.
Seymour, M., I. Durance, B.J. Cosby, et al. 2018. Acidity promotes degradation of multi-species environmental DNA in lotic mesocosms. Communications Biology 1(4). https://doi.org/10.1038/s42003-017-0005-3.
Shelton, A.O., R.P. Kelly, J.L. O’Donnell, et al. 2019. Environmental DNA provides quantitative estimates of a threatened salmon species. Biological Conservation 237: 383–391. https://doi.org/10.1016/j.biocon.2019.07.003.
Shogren, A.J., J.L. Tank, E. Andruszkiewicz, et al. 2017. Controls on eDNA movement in streams: Transport, retention, and resuspension. Scientific Reports 7(5065). https://doi.org/10.1038/s41598-017-05223-1.
Shogren, A.J., J.L. Tank, S.P. Egan, et al. 2018. Water flow and biofilm cover influence environmental DNA detection in recirculating streams. Environmental Science and Technology 52: 8530–8537. https://doi.org/10.1021/acs.est.8b01822.
Simberloff, D. 2021. Maintenance management and eradication of established aquatic invaders. Hydrobiologia 848: 2399–2420. https://doi.org/10.1007/s10750-020-04352-5.
Smith, K.F., S.A. Wood, D.O. Mountfort, and S.C. Cary. 2012. Development of a real-time PCR assay for the detection of the invasive clam, Corbula amurensis, in environmental samples. Journal of Experimental Marine Biology and Ecology 412: 52–57. https://doi.org/10.1016/j.jembe.2011.10.021.
Sommer, T., C. Armor, R. Baxter, et al. 2007. The collapse of pelagic fishes in the Upper San Francisco Estuary. Fisheries 32: 270–277. https://doi.org/10.1577/1548-8446(2007)32.
Spear, M.J., H.S. Embke, P.J. Krysan, and M.J. Vander Zanden. 2021. Application of eDNA as a tool for assessing fish population abundance. Environmental DNA 3(1): 83–91. https://doi.org/10.1002/edn3.94.
Stat, M., M.J. Huggett, R. Bernasconi, et al. 2017. Ecosystem biomonitoring with eDNA: Metabarcoding across the tree of life in a tropical marine environment. Scientific Reports 7(12240). https://doi.org/10.1038/s41598-017-12501-5.
Stewart, K.A. 2019. Understanding the effects of biotic and abiotic factors on sources of aquatic environmental DNA. Biodiversity and Conservation 28: 983–1001. https://doi.org/10.1007/s10531-019-01709-8.
Stoeckle, M.Y., M. Das Mishu, and Z. Charlop-Powers. 2020. Improved Environmental DNA reference library detects overlooked marine fishes in New Jersey United States. Frontier in Marine Science 7. https://doi.org/10.3389/fmars.2020.00226.
Stoeckle, M.Y., L. Soboleva, and Z. Charlop-Powers. 2017. Aquatic environmental DNA detects seasonal fish abundance and habitat preference in an urban estuary. PLoS One 12(4): e0175186. https://doi.org/10.1371/journal.pone.0175186.
Stompe, D., P. Moyle, A. Kruger, and J. Durand. 2020. Comparing and integrating fish surveys in the San Francisco Estuary : Why diverse long-term monitoring programs are important. San Francisco Estuary and Watershed Science 18. https://doi.org/10.15447/sfews.2020v18iss2art4.
Strickler, K.M., A.K. Fremier, and C.S. Goldberg. 2015. Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biological Conservation 183: 85–92. https://doi.org/10.1016/j.biocon.2014.11.038.
Taberlet, P., A. Bonin, L. Zinger, and E. Coissac. 2018. Environmental DNA for biodiversity research and monitoring. Oxford, UK: Oxford University Press.
Takasaki, K., H. Aihara, T. Imanaka, et al. 2021. Water pre-filtration methods to improve environmental DNA detection by real-time PCR and metabarcoding. PLoS One 16(9): e0258073. https://doi.org/10.1371/journal.pone.0250162.
Teffer, A.K., J. Carr, A. Tabata, et al. 2020. A molecular assessment of infectious agents carried by Atlantic salmon at sea and in three eastern Canadian rivers, including aquaculture escapees and North American and European origin wild stocks. Facets 5: 234–263. https://doi.org/10.1139/facets-2019-0048.
Teffer, A.K., S. Hinch, K. Miller, et al. 2019. Cumulative effects of thermal and fisheries stressors reveal sex-specific effects on infection development and early mortality of adult Coho salmon (Oncorhynchus kisutch). Physiological and Biochemical Zoology 92: 505–529. https://doi.org/10.1086/705125.
Thalinger, B., K. Deiner, L.R. Harper, et al. 2021. A validation scale to determine the readiness of environmental DNA assays for routine species monitoring. Environmental DNA 3(4): 823–836. https://doi.org/10.1101/2020a.04.27.063990.
Thalinger, B., D. Kirschner, Y. Pütz, et al. 2020. Lateral and longitudinal fish environmental DNA distribution in dynamic riverine habitats. Environment DNA 3(1): 305–318. https://doi.org/10.1002/edn3.171.
Thomas, A. C., S. Tank, P. L. Nguyen, et al. 2020. A system for rapid eDNA detection of aquatic invasive species. Environmental DNA 2(3): 261–270. https://doi.org/10.1002/edn3.25.
Thomsen, P.F., and E. Willerslev. 2015. Environmental DNA – An emerging tool in conservation for monitoring past and present biodiversity. Biological Conservation 183: 4–18. https://doi.org/10.1016/j.biocon.2014.11.019.
Tillotson, M. D., R.P. Kelly, J. J. Duda, et al. 2018. Concentrations of environmental DNA (eDNA) reflect spawning salmon abundance at fine spatial and temporal scales. Biological Conservation 220: 1–11. https://doi.org/10.1016/j.biocon.2018.01.030.
Tingley, R., R. Coleman, N. Gecse, et al. 2021. Accounting for false positive detections in occupancy studies based on environmental DNA: A case study of a threatened freshwater fish (Galaxiella pusilla). Environmental DNA 3(2): 388–397. https://doi.org/10.1002/edn3.124.
Tsuji, S., T. Takahara, H. Doi, et al. 2019. The detection of aquatic macroorganisms using environmental DNA analysis–A review of methods for collection, extraction, and detection. Environmental DNA 1(2): 99–108. https://doi.org/10.1002/edn3.21.
Tsuri, K., S. Ikeda, T. Hirohara, et al. 2021. Messenger RNA typing of environmental RNA (eRNA): A case study on zebrafish tank water with perspectives for the future development of eRNA analysis on aquatic vertebrates. Environmental DNA 3(1): 14–21. https://doi.org/10.1002/edn3.169.
Turner, C.R., M.A. Barnes, C.C.Y. Xu, et al. 2014. Particle size distribution and optimal capture of aqueous macrobial eDNA. Methods in Ecology and Evolution 5: 676–684. https://doi.org/10.1111/2041-210X.12206.
Turner, C.R., K.L. Uy, and R.C. Everhart. 2015. Fish environmental DNA is more concentrated in aquatic sediments than surface water. Biological Conservation 183: 93–102. https://doi.org/10.1016/j.biocon.2014.11.017.
von Ammon, U., S.A. Wood, O. Laroche, et al. 2019. Linking environmental DNA and RNA for improved detection of the marine invasive fanworm Sabella spallanzanii. Frontiers in Marine Science 6: 1–13. https://doi.org/10.3389/fmars.2019.00621.
Welsh, A., C. Jerde, C. Wilson, et al. 2020. Management support tree for the interpretation of positive laboratory results. Great Lakes Fishery Commission.
Wilcox, T.M., K.S. McKelvey, M.K. Young, et al. 2015. Environmental DNA particle size distribution from Brook Trout (Salvelinus fontinalis). Conservation Genetics Resources 7: 639–641. https://doi.org/10.1007/s12686-015-0465-z.
Williams, M., C. Hernandez, A.M. O’Sullivan, et al. 2021. Comparing CRISPR-Cas and qPCR eDNA assays for the detection of Atlantic salmon (Salmo salar L.). Environmental DNA 3(1): 297–304. https://doi.org/10.1002/edn3.174.
Wood, S.A., X. Pochon, O. Laroche, et al. 2019. A comparison of droplet digital polymerase chain reaction (PCR), quantitative PCR and metabarcoding for species-specific detection in environmental DNA. Molecular Ecology Resources 19: 1407–1419. https://doi.org/10.1111/1755-0998.13055.
Yates, M.C., A.M. Derry, and M.E. Cristescu. 2021. Environmental RNA: A revolution in ecological resolution? Trends in Ecology & Evolution 36: 601–609. https://doi.org/10.1016/j.tree.2021.03.001.
Yates, M.C., D.J. Fraser, and A.M. Derry. 2019. Meta-analysis supports further refinement of eDNA for monitoring aquatic species-specific abundance in nature. Environmental DNA 1(1): 5–13. https://doi.org/10.1002/edn3.7.
Zou, K., J. Chen, H. Ruan, et al. 2020. eDNA metabarcoding as a promising conservation tool for monitoring fish diversity in a coastal wetland of the Pearl River Estuary compared to bottom trawling. Science of the Total Environment 702: 134704. https://doi.org/10.1016/j.scitotenv.2019.134704.
Acknowledgements
We thank the Delta Stewardship Council for funding the publication of this paper and partially funding the symposium. We thank Carole Hom for leading the organization of the symposium and UC Davis Coastal and Marine Sciences Institute (CMSI) for providing funding for the symposium. We thank Brian Schreier, Ted Sommer, and Nicole Kwan for helpful comments and suggestions, and Sarah Stinson for help with notetaking at the post-symposium round table discussion. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the United States Government.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Kenneth L. Heck
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nagarajan, R.P., Bedwell, M., Holmes, A.E. et al. Environmental DNA Methods for Ecological Monitoring and Biodiversity Assessment in Estuaries. Estuaries and Coasts 45, 2254–2273 (2022). https://doi.org/10.1007/s12237-022-01080-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12237-022-01080-y