Abstract
Potato market value is heavily affected by tuber quality traits such as shape, color, and skinning. Despite this, potato breeders often rely on subjective scales that fail to precisely define phenotypes. Individual human evaluators and the environments in which ratings are taken can bias visual quality ratings. Collecting quality trait data using machine vision allows for precise measurements that will remain reliable between evaluators and breeding programs. Here we present TubAR (Tuber Analysis in R), an image analysis program designed to collect data for multiple tuber quality traits at low cost to breeders. To assess the efficacy of TubAR in comparison to visual scales, red-skinned potatoes were evaluated using both methods. Broad sense heritability was consistently higher for skinning, roundness, and length to width ratio using TubAR. TubAR collects essential data on fresh market potato breeding populations while maintaining efficiency by measuring multiple traits through one phenotyping protocol.
Resumen
El valor de mercado de la papa se ve muy afectado por los rasgos de calidad del tubérculo, como la forma, el color y el despellejamiento. A pesar de esto, los mejoradores de papa a menudo se basan en escalas subjetivas que no definen con precisión los fenotipos. Los evaluadores humanos individuales y los entornos en los que se toman las calificaciones pueden sesgar las calificaciones de calidad visual. La recopilación de datos de rasgos de calidad mediante visión artificial permite mediciones precisas que seguirán siendo confiables entre los evaluadores y los programas de mejoramiento. Aquí presentamos TubAR (Tuber Analysis in R), un programa de análisis de imágenes diseñado para recopilar datos para múltiples rasgos de calidad de tubérculos a bajo costo para los mejoradores. Para evaluar la eficacia de TubAR en comparación con las escalas visuales, se evaluaron las papas de piel roja utilizando ambos métodos. La heredabilidad de sentido amplio fue consistentemente mayor para el despellejamiento, la redondez y la relación longitud / anchura usando TubAR. TubAR recopila datos esenciales sobre las poblaciones de reproducción de papa fresca del mercado mientras mantiene la eficiencia al medir múltiples rasgos a través de un protocolo de fenotipado.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tuber quality traits, including shape, color, and skinning (excoriation), are key determinants for the market value of potatoes (Carputo et al. 2004). Therefore, they are important selection targets for potato breeders. These tuber quality traits are traditionally rated on subjective five or nine point visual scales (Reeves 1988; Van Eck et al. 1994; Prashar et al. 2014; Buhrig et al. 2015). While these scales simplify breeder note taking and allow for comparison across programs, they may not encompass all meaningful aspects of a trait. For instance, in the US National Chip Processing Trial, tuber shape is rated on a five-point scale from compressed to elongate as shown in Fig. 1. The scale fails to account for other deviations from ovoid, such as shoulders/or a more rectangular shape, pointy ends, or lumpiness. Use of calipers to measure tuber length to width ratios can provide a more precise measurement of tuber shape; however, this is labor intensive and does not capture other elements of tuber shape that are important to processors and consumers, such as second growth which can lead to loss of yield in harvesting and processing (Chung et al. 1988).
USDA form OMB NO 0581 − 0055 Tuber shape classification. (USDA Plant Variety Protection Office 2015). This is converted into a numerical scale by breeding programs generally 1 (compressed) to 5 (long)
Several sources of inconsistency arise in applying visual ratings. Results can vary across scientists due to individual differences in sensory perception (e.g. varying degrees of color perception capacity) and internalized definition of the target phenotype, as well as across time due to breeder experience level, fatigue, and differences in lighting during evaluation (Parker et al. 1995; Poland and Nelson 2011). In research involving many scientists over a large area, consistent, clear phenotyping standards are vital to ensuring high quality data and increasing the likelihood of meaningful results (Parker et al. 1995; Poland and Nelson 2011). Within-field or within-experiment check clones may be used to “visually calibrate” one’s scoring before evaluating experimental entries. However, genotype by environmental interactions account for a portion of phenotypic variance and relying on generalizations about the standard cultivars’ expected performance would result in a visual mis-calibration in some instances (Liu et al. 2019). Machine vision can improve consistency across programs and environments by eliminating differences due to evaluator bias, as well as measure traits of interest more accurately (Parker et al. 1995; Bock et al. 2008; Poland and Nelson 2011).
Machine vision is commonly utilized to assist in commercial quality sorting in a variety of crops including potato. Most machine vision platforms for potatoes are designed for industrial scale processors and implementing these systems in breeding programs may involve expensive equipment and may not provide the precision or consistency needed for research applications (Cubero et al. 2011; Moreda et al. 2012; Zhou et al. 1998). Specialized equipment for measurement of color (colorimeter) and skinning (torque meter) exist, but these are expensive and labor intensive as they are limited to the evaluation of one or very few traits (Krupek et al. 2021; Roe et al. 2014). Accurate and precise image analysis of multiple tuber traits using standard digital camera images would therefore, be advantageous to efficient and low-cost measurement of quantitative tuber quality traits (Si et al. 2017; Caraza-Harter and Endelman, 2020; Neilson et al. 2021).
Image analysis systems for potato tubers with accuracy appropriate for research and breeding program applications have been previously developed for both tuber shape and color. Si et al. (2017) used watershed segmentation to identify tubers in standard two-dimensional digital images and measured the length to width ratio of the segmented tuber shapes. Accuracy of the ratios compared to caliper measurements was 96% for white-skinned potatoes and 94% for red-skinned potatoes, with the lower accuracy in red potatoes attributed to their generally more circular shape. Neilson et al. (2021) developed aspect ratio and circularity measures to account for the aspects of shape that cannot be accounted for by length to width ratio. Caraza-Harter and Endelman (2020) evaluated red skin color and skinning in digital images using the RGB Measure Plugin in ImageJ (Schneider et al. 2012). Plot based heritability, using image analysis for phenotyping, was above 0.75 for the hue, chroma, and lightness measures of skin color, as well as for skinning.
While these image analysis methods provide accurate results at a low cost, high throughput phenotyping requires more automation and simplification of the analysis process. The tuber image analysis process could greatly benefit from having a single program and protocol to measure multiple quality traits at once. TubAR (Tuber Analysis in R) provides a simple and efficient method for phenotyping multiple tuber traits from light box images simultaneously.
Materials and Methods
Package Development
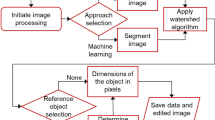
We have developed an R package called TubAR (Tuber Analysis in R) which calculates values for quality traits from lightbox images. Currently it measures roundness, length to width ratio, lightness, redness, and skinning. We describe the calculations and steps for analysis within the pipeline in the next four sections and then the images and protocols used for validation and testing.
Image Segmentation
Tubers were isolated from the image background through image segmentation. Images were read into the R statistical software (R Core Team 2018) environment with the package EBImage (Pau et al. 2010) and resized to reduce computation time with the function resize. Image size was reduced by a factor of 4 from 5840 × 3940 to 1460 × 985. Transformation from RGB to the CIELab color scale was done with the function convertColor (R Core Team 2018). We chose the CIELab color scale because it is the international standard for description of the color of objects due to its similarity to human perception (International Commission on Illumination 2019). Each component matrix of the color scales (R, G, B, L, a, and b) was inspected for capacity to create separation between tuber and background pixels. The resulting components of the image were subjected to a threshold to create a binary filter. Within-tuber gaps, due to lightly colored patches of skin or skinning, were filled with the function fillHull (Pau et al. 2010). Image segmentation was conducted with the function bwlabel (Pau et al. 2010). A minimum object size of 4000 pixels was enforced to remove objects resulting from shadows and small debris (e.g. tuber skin flakes and residual soil).
Color Correction
We used three-dimensional thin-plate spline (TPS) to color correct images to minimize the effects of any changes in light levels. TPS for color correction warps (transforms) pixel color values of a conserved object in an image to a predetermined reference RGB color value, in this case, the center pixel of each color chip in the 24-chip color card. An interpolation function is then used to transform the color values of the other pixels in the image using the difference between the observed color of the color card pixels and the reference colors (Menesatti et al. 2012). The tps3d function from the Morpho package (Schlager 2017) was utilized to create the interpolation function and determine the corrected color values for each pixel.
Shape Measures
Several measures were calculated on a per-tuber basis with the objective of identifying measures that correlate well with visually apparent aspects of tuber shape and identify deviations from ideal shapes. The perimeter and area were recorded for each tuber, as well as the convex hull of each tuber. The convex hull is the tuber shape modified by removing indents, which are often created by micro-environmental factors. Maximum length was determined as the maximum of distances between all possible pairs of perimeter pixels. The minimum bounding box was fit for each tuber resulting in measures of length, width, and length to width ratio. Convex hull area and convex hull perimeter were used to calculate roundness. Roundness ranges from zero (for a straight line) to one (for a perfect circle) (Van der Werff and Van der Meer 2008).
Color and Skinning Measures
On a per tuber basis, pixels were converted from RGB to the CIELab color scale with the function convertColor. We used the nlsLM function from the minpack.lm package (Elzhov et al. 2016) to fit a sigmoid curve with the formula:
in which y is the percent of the tuber pixels designated skinned at b (from Lab) threshold x. The parameters a, c, d, and e were initiated at the maximum y value, 1, the median x value, and minimum y value, respectively. The threshold for designating pixels skin (red) or skinned (flesh, white to yellow) was set at 1.5x the d value, whereby the algorithm responds to differences in tuber skin and flesh colors for each image. It is assumed that all tubers within an image comprise a sample from the same variety under the same treatment.
Excluding the portions identified as skinned, tuber skin color was quantified in terms of redness as the median a (from Lab) value for each tuber and skin lightness as the median L (from Lab) value for each tuber (Fig. 2).
Plant Material and Phenotyping
We collected a random sample of 10 USDA medium (6.35–8.255 cm) sized potatoes from each plot of a nitrogen trial with four replicates conducted at the Sand Plain Research Farm in Becker, MN in the summers of 2018 and 2019 described in Stefaniak et al. (2021). Entries were advanced red potato selections from the University of Minnesota breeding program and a selection of red potato cultivars favored by Minnesota growers. All entries were planted in 20 hill plots replicated four times per year. Plots were desiccated 90 days after planting (DAP) using diquat and mechanically harvested 104 DAP; tubers were sized with a Kerian sizer (Kerian Machines Inc. Grafton, ND). In 2018, plots were graded in a three day period three weeks after harvest, while in 2019 plots were graded in a three day period directly following harvest. Tuber shape was rated categorically as “rectangular”, “elongate”, “oblong”, “pear” or “round”. Skinning was rated on a 0 to 5 scale (none to severe), and tuber color intensity was rated on a 0 to 5 scale (white to intense red color).
Additionally, we collected samples of red potato clones from the University of Minnesota breeding program that were grown over the 2019 and 2020 field seasons. In 2019 potatoes were grown at the North Central Research and Outreach Center in Grand Rapids, MN in single replicate 12-hill plots. Plots were desiccated 98 DAP and mechanically harvested 118 DAP. These tubers were not graded or rated using visual scales. In 2020 the same clones were grown at the Sand Plain Research Site in single replicate 20 hill plots. Plots were desiccated 90 DAP and mechanically harvested 104 DAP. In 2020 tubers were graded on an AgRay sorter (Agray Vision Systems, Acampo, CA) and visually rated as above. They were graded within the three day period immediately following harvest. All visual ratings across the three years were made by Dr. Shannon.
Following the methods described by Caraza-Harter and Endelman (2020), tubers were gently washed with water to remove soil and allowed to dry. Although we did not maintain a consistent drying time, tubers were photographed after they were no longer damp. For each plot, ten tubers were staged in an Ortery Photosimilie 200 software-controlled light box with a 6500 K daylight illumination system (Ortery Technologies Inc.) with front and rear lights turned on. One image was taken per plot. Photographs were taken with a Rebel T6i camera with a 24 mm lens, ISO 100, 1/30 sec shutter speed and aperture f/5.6. A CameraTrax 24ColorCard (CameraTrax.com CT24-23-1315) was placed in the lower right corner of the image. Images were 6000 × 4000 pixels with resolution of 72 × 72. Images were saved in .jpg (lossy compression) format. Photographs were taken over the month following grading.
Heritability Calculations
Broad sense heritability for redness and skinning was calculated using data from visual ratings and then from image analysis data collected from the potato tubers. We used the lmer function from the lme4 R package (Bates et al. 2015) to create linear mixed-effect models from trait values, clone, nitrogen rate, and block among the nitrogen trial populations. Separate models were calculated for each year.
Where Pijk is the trait phenotype of clone i in block j at nitrogen rate k, µ is the intercept, Gi is the random effect of genotype, Bj is the effect of block, Rk is the effect of nitrogen rate, and εijk is the random effect of the residual. For the breeding program data, field year was used instead of block and no effect from nitrogen was present, making the linear mixed-effect model.
Variance components, VG and VE, were taken from the lmer function output and used to estimate broad sense heritability.
Sample Size Variance Calculations
In order to evaluate the optimal number of tubers per photo, we determined the mean standard error of standardized output values for each of the five phenotypes described above using 3 to 10 tuber samples, which is the practical limit of what can easily fit in the lightbox for most clones. Samples were taken from images of 224 clones harvested from the University of Minnesota breeding program in 2020. Smaller sample sizes were simulated from trait data of 10 samples by shortening the sample data lists. Because tuber placement within images was not ordered, these smaller tuber samples were still random. For each trait, the mean was standardized with respect to units so all traits could be plotted together and the standard error at a given tuber number was averaged across the 224 clones. These averages were plotted to evaluate gain in precision from each additional tuber included in an image.
Visual Scale Consistency Test
We created a set of 50 red potato sample pictures, then created two more sets of the same sample pictures in randomized orders. All tubers had red skin and white flesh. The skin.all function in TubAR was used to remove the backgrounds of the photos to limit information about the clone genotype. Four members of the University of Minnesota Potato Breeding and Genetics Lab with prior experience in rating tubers using visual scales rated the pictures from each of the three sets using the skinning, color intensity, and shape visual scales previously discussed. Scores were then derandomized and the coefficient of determination (R2) within the sets scored by the same evaluator, as well as between each evaluator, was determined using the cor function in R. The maximum, minimum, and average coefficient of determination was determined for scores of different sets from the same evaluator as well as between different evaluators.
Because the skinning trait rated by TubAR is theoretically comparable to that rated with the visual scale, the same set of 50 sample pictures was rated for skinning using TubAR. The ratings were converted from percentages to a five point scale, mimicking the ratings from human observers. This set of skinning values was compared to each of the sets rated by the human evaluators using the maximum, minimum, and average coefficient of determination as discussed above. The standard deviation for visual scores of each trait for each sample image were averaged for each trait to measure variation in rating across evaluators.
An additional set of 30 sample images of red potatoes collected from the USDA-ARS Potato Breeding and Genetics program in Prosser, WA, using a Nikon D7100 DSLR camera and a HAVOX HPB-80D photo studio light box with a non-reflective black background. These images were taken after at least 90 days in storage. Images were evaluated by a scientist from the USDA and one from the University of Minnesota to determine difference in ratings between programs and demonstrate TubAR’s extendibility to other lightbox set ups. Color intensity and shape were rated, while skinning was not due to low variance among the sample images. Trait scores were compared within one evaluator’s scores and between evaluators based on R2 values. Human color ratings were compared to lightness and redness values from TubAR and human shape scores were compared to length to width ratio and roundness using R2 values.
Time Efficacy Test
To quantify the difference in time to get trait data using TubAR versus previously available methods for quantitative tuber trait measurement, we timed the use of TubAR, calipers, and a colorimeter to collect data from 10 tubers.
For TubAR measurements, timing was started at the beginning of staging the tubers in the lightbox. Once a photo of 10 tubers was taken and saved, the timer was stopped. An R script was run on a personal computer (AMD Ryzen 5 3600 3.6 GHz processor, 16 GB of RAM, Microsoft Windows 10 Pro version 21H1) to collect skin and shape data from the photo, with the timer being restarted upon running the script, and stopped at the console printing trait values. To reflect the scalability of TubAR, a sample of 100 photos was processed with TubAR using the skin.all function using four processor cores, timing started upon running the R script and ended upon the creation of matrices of median skin and shape trait values.
For the alternative manual measurement method, ten tubers, calipers, a reflectance colorimeter (Photovolt Instruments Photoreflectometer 577PC), and a computer were assembled beforehand, and a timer was started once measurement began. Length to width measurements and two values of percent reflectance were taken for each tuber and recorded in a spreadsheet, the timer was stopped after the last value was recorded. This was performed by three individuals and the average time was used in further calculations.
Total time to record data for one sample was directly timed for both methods, while time to collect data for 100 samples was based on multiplying the time to perform the manual component of each method, and in the case of TubAR adding the processing time for 100 photos.
Weight Area Correlation
A total of 337 potato breeding lines and named cultivars maintained by the USDA-ARS Potato Breeding and Genetics program in Prosser, WA, and the U.S. Potato Genebank at Sturgeon Bay, WI, were planted as two replicates of five-hill plots in the Pear Acres field site at Washington State University Irrigated Agricultural Research and Extension Center at Prosser, WA. Tubers from the plots were assessed for yield in order to compare tuber weight to tuber area. Tubers were rinsed by applying water to the samples for several minutes inside modified Kobalt 4-cu ft 0.5-HP cement mixers (Lowes LLC) lined with a ¼ inch yoga mat material (YogaDirect.com LLC). Measurements of sample yield were be taken using Ohaus Valor 7000 scale controlled by Python script run on a Raspberry Pi 3 computer. Those same samples were photographed and total tuber area in pixels was determined for each sample using TubAR. The cor function in R was used to determine the R2 between the weight data and total area data from each sample.
Results
Parameters for TubAR Use
Image size was reduced by a factor of four which was found to speed computation time while preserving the image sufficiently, as determined by visual inspection of the reduced images. Examples of the original and reduced images are shown in Fig. 3.
To determine how the number of tubers per image affects the consistency of the measurements, we calculated standard errors for subsets of the tubers in each image. For all traits we observed a decrease in standard error with each additional tuber. However, the marginal effect of each additional tuber decreased as total tuber number increased (Fig. 4).
Effectiveness as Compared to Visual Ratings
We compared the performance of TubAR to visual ratings by looking at heritabilities and correlations. TubAR trait heritabilities were consistently higher than visual scores for the 2018 population (Table 1). In the 2019 nitrogen trial visual color intensity heritability was higher, with TubAR redness and lightness heritabilities being comparatively lower. TubAR trait heritabilities were recorded for the breeding program populations but could not be compared to visual scores because they were only scored visually one year. We could not calculate heritability for skinning in the breeding population due to low variation across clones.
R-squared values between comparable visual and TubAR measurements ranged from 0.26 to 0.41 (Fig. 5). We observed negative correlations for roundness and lightness because the scales for the TubAR measurements and the visual measurements run in opposite directions.
Scatterplots and regression lines of a Skinning percent determined by TubAR vs. skinning 1 to 5 visual rating, b Redness from TubAR vs. color intensity 1 to 5 visual rating, c Lightness from TubAR vs. color intensity 1 to 5 visual rating, (while 100 is the lightest TubAR value, 1 is the lightest visual score) d Roundness as measured by TubAR vs. shape visual rating (while 1 is the roundest TubAR value, 1 is the roundest visual rating), e Length to width Ratio taken from the length and width measured by TubAR vs. shape visual rating
Judging the effectiveness of TubAR using correlations assumes the visual ratings are accurate. We had multiple evaluators rate the same images to determine consistency across evaluators. Coefficients of determination for human visual scores (Table 2) tended to be higher between ratings by the same individual than between individuals but ranged widely in every case (less than 0.2 to more than 0.6). Shape scores had the highest within and between correlations while color intensity had the lowest. The average R2 between TubAR scores and human ratings was very similar to the between evaluator average (≈ 0.25). The average standard deviation of scores across the ratings for color intensity was 0.71, 0.76 for skinning, and 0.39 for shape.
In order to look at evaluator consistency in a different population and between different breeding programs, we also had a evaluator from the University of Minnesota and the USDA rate tubers from the USDA for color intensity and shape. Within evaluator R2 values averaged 0.54 for color and 0.66 for shape. Between evaluator R2 values averaged 0.12 for color and 0.60 for shape. TubAR values for lightness and redness were compared to color intensity. Length to width ratio and roundness were compared to shape. Color intensity had an average R2 of 0.11 with redness and 0.41 with lightness. Shape rating had an average R2 of 0.58 with length to width ratio and 0.65 with roundness.
Comparing tuber weight and tuber area as calculated by TubAR provides a measure of effectiveness that does not depend on the accuracy of visual ratings. The R2 between tuber sample weights and total tuber photo area was 0.77.
Time Investment
Measuring and recording color and length to width ratio for a single, ten-tuber sample took 5 min 40 s using TubAR, and 4 min 14 s using a calipers and photovolt on each tuber. Taking a picture for TubAR took 2 min 23 s with the remaining time being processing in R. Processing 100 samples in TubAR took 54 min 1 s. When the photo taking time is multiplied by 100 and added to TubAR processing time, the expected time to collect data for 100 samples in TubAR is 4 h 52 min. The caliper and colorimeter time is multiplied by 100 to get the expected manual measurement time of 7 h 3 min. Washing tubers before photography would add to the amount of time required for TubAR.
Discussion
TubAR was created to improve the precision and accuracy of tuber quality trait measurements over that of the visual scales often used by breeding programs to phenotype for selection. The more accurate the information used in breeder decisions, the more effective selection can be. This is particularly important as potato breeders begin implementing genomic selection cycles, where inaccurate predictions can drive populations in the wrong direction (Caruana et al. 2019; Enciso-Rodriguez et al. 2018; Endelman et al. 2018; Gemenet et al. 2020; Habyarimana et al. 2017; Selga et al. 2021; Sood et al. 2020; Stich and Van Inghelandt 2018; Sverrisdóttir et al. 2017; Sverrisdóttir et al. 2018).
Accuracy and Precision
When visual ratings are used, clones are assigned a whole number value between one and five. TubAR increases the potential for precision by relying on a continuous numeric scale rather than a rating. Accuracy is more difficult to measure, but from a breeding perspective, it is most important to be accurate about the genetic component of a trait. For traits measured using multiple methods in the same population, comparing heritability can provide information on the relative accuracy of measurement techniques (Caraza-Harter and Endelman 2020). Because location and genotype remain the same across measures, any changes in heritability can be attributed to the accuracy of the phenotyping method. TubAR roundness, length to width ratio, and skinning consistently had higher heritability than the visual scale measurements, while redness, and lightness did not (Table 1).
It is important to note that redness and lightness are affected by tuber washing/drying. Visual inspection showed that images from 2018, where both TubAR color measurements exhibited high heritability as compared to the visual scores, featured consistently dry tubers, whereas images from 2019, where heritability was dramatically lower, featured some wet and some dry tubers. Washing tubers before taking pictures improves the accuracy of color ratings but all tubers should be left to completely dry before being photographed. We recommend letting tubers dry at least 24 h, however the details of the image capture protocol are less important than the consistency of that protocol. TubAR is only as good as the input images, therefore care must be taken to produce consistent images.
The increases we observed in heritability for many TubAR traits in comparison to visual scales could be attributed to either an improvement in our ability to measure traits or to refining the definition of the traits. Breaking down a trait like shape or color into its component pieces may better reflect the underlying factors. For example, lightness increases with length of time in storage, while hue, or color family, remains unchanged (Caraza-Harter and Endelman 2020). Similarly, nitrogen affects lightness but not redness (Jones et al. 2021) and heat affects the expression levels of anthocyanins but not which ones are expressed (Liu et al. 2019). Separating color into lightness and redness, may be allowing us to distinguish between the aspects of color more dependent on environment and those that are more heavily genetic.
A second method we explored for determining the accuracy of TubAR measurements was comparing the values to visual scores (Fig. 5). However, the correlation between scores from different evaluators (Table 2) were so low as to bring into question the value of the visual ratings as a benchmark. The correlation between TubAR traits and visual ratings was consistent with the correlation between measurements from different evaluators. We observed a higher correlation when comparing area as determined from images to tuber weight. However, this correlation was lower than the extremely high correlation (R2 = 0.94) reported by Zhou et al. (1998) and Neilson et al. (2021). The discrepancy could be accounted for by the fact that Zhou et al. only examined three potato cultivars and Neilson et al. focused on crosses for processing while we measured diverse samples from a fresh market breeding program where the requirements for specific gravity are less stringent. Significant variation in area to weight correlation between cultivars has been found in mango (Patel et al. 2020). This is likely to be true for potatoes as well, given genetic variation in shape and specific gravity both of which would influence the correlation between weight and area (Slater et al. 2014). We do not recommend using TubAR to estimate tuber weight.
Practicality of Implementation
We were able to collect and process images for a very low material cost. While some labor cost is saved in the faster processing of many images, staging and taking photos of each set of tubers is still labor intensive. However, when taking measurements for many samples, time is ultimately saved compared to using calipers and a colorimeter to manually measure traits. Additionally, TubAR measures more traits than the two measured by manual devices and adding trait measurements does not add to the time it takes to collect data. It should also be noted that much of the processing time using TubAR does not require a human to be actively involved, potentially lowering labor costs well beyond the time saved compared to manual measurement.
The time required in staging tubers can be balanced with the precision desired in an experiment. The standard errors seen in different sample sizes largely decrease with every additional tuber from three up to the maximum ten in each trait. While the standard errors continue to decrease, each additional tuber decreases the standard error less than the previous tuber. This creates a situation of diminishing returns where the gain in precision will eventually not make up for the extra time spent staging more tubers. This tradeoff is particularly important for larger market classes, such as russets, where fitting ten tubers in one image may not be practical for a lightbox, or when time and labor available for phenotyping is constrained.
All measurements with calipers, photovolt, and lightbox were taken by experienced practitioners. As with any lab procedure, each technique has a learning curve which is not factored into the time estimates above. The learning curve for the TubAR system is not steep. Undergraduates in the Shannon lab regularly use the lightbox and analyze the resulting images.
Potential Benefits
Machine vision phenotyping systems such as TubAR can provide more accuracy from year to year within a breeding program if the same system is used to evaluate clones from generation to generation. They can also be used to increase accuracy between programs in situations such as national trials where multiple evaluators would introduce bias to visual scales, as seen by the low correlations between ratings of different scientists.
Machine vision has been implemented in other crops. Most commonly it is used for quality sorting in commercial settings. Within research settings, image analysis has been used to diagnose disease in cereals (Parker et al. 1995), maize (Poland and Nelson 2011), and grapefruit (Bock et al. 2008). One of the most widely used tools for crop image analysis is Plant CV (Gehan et al. 2017). Plant CV aims to be broadly useful for a variety of tissues and image types (RGB, NIR, hyperspectral etc.). The tradeoff for applicability to a variety of crops and situations and extensive user control is that it takes more effort and expertise to implement.
While TubAR’s ability to score many traits from one image increases the speed that each of those traits can be scored compared to manual or visual scale measurements, staging tubers, tags, and color cards for each photo is still labor intensive and sets a practical limit on the amount of tubers that could be measured in a year by most breeding or research programs. Modifying this program for use in a potato sorter may allow high-throughput phenotyping by eliminating the need to manually stage each photograph. Additionally, certain sorters take pictures from multiple angles allowing length to thickness to be measured as well as length to width ratio.
Our current version of TubAR focuses on quality traits crucial to fresh market red potato including skin color, shape, and skinning. There is potential for other traits of interest to be measured with TubAR through a similar image analysis which would increase the applicability of TubAR to other market classes. Traits of interest that may include russeting, greening, eye depth, and diseases including common scab, Rhizoctonia, and silver scurf. Although some of these traits can likely be measured with thresholds and measurements similar to those used above, many traits essential to tuber quality are more complicated and will require a machine learning approach. One benefit of TuBAR is that we can retain images and as we add functionality, we can measure additional traits in previous years’ breeding populations. Only limited modifications may be necessary to use the program to measure the color of chips and fries or even the color and shape of other crops such as apples.
TubAR is available at https://github.com/shannonlabumn/TubAR. Instructions, a vignette, and sample image data are available for download with the package. Source code can also be found in the github repository.
References
Bates, D., M. Maechler, B. Bolker, and Walker, S. 2015. Fitting linear mixed-effects models using lme4. Journal of Statistical Software 67:1–48.
Bock, C.H., P.E. Parker, A.Z. Cook, and T.R. Gottwald. 2008. Visual rating and the use of image analysis for assessing different symptoms of citrus canker on grapefruit leaves. Plant Disease 92: 530–541.
Buhrig, W., M.K. Thornton, N. Olsen, D. Morishita, and C. McIntosh. 2015. The influence of ethephon application timing and rate on plant growth, yield, tuber size distribution and skin color of Red LaSoda potatoes. American Journal of Potato Research 92: 100–108.
Caraza-Harter, M.V., and J.B. Endelman. 2020. Image‐based phenotyping and genetic analysis of potato skin set and color. Crop Science 60: 202–210.
Carputo, D., R. Aversano, and L. Frusciante. 2004. Breeding potato for quality traits. Meeting of the Physiology Section of the European Association for Potato Research 684: 55–64.
Caruana, B.M., L.W. Pembleton, F. Constable, B. Rodoni, A.T. Slater, and N.O.I. Cogan. 2019. Validation of genotyping by sequencing using transcriptomics for diversity and application of genomic selection in tetraploid potato. Frontiers in Plant Science 10: 670. https://doi.org/10.3389/fpls.2019.00670.
Chung, B., D. Armstrong, and S. Grice. 1988. Effect of irrigation frequency on the incidence of second growth of Russet Burbank potatoes in north-west Tasmania. The Journal of Agricultural Science 111: 399–402.
Cubero, S., N. Aleixos, E. Moltó, J. Gómez-Sanchis, and J. Blasco. 2011. Advances in machine vision applications for automatic inspection and quality evaluation of fruits and vegetables. Food and Bioprocess Technology 4: 487–504.
Elzhov, T. V., K. M. Mullen, A. N. Spiess, and B. Bolker. 2016. Minpack.lm: R interface to the Levenberg-Marquardt nonlinear least-squares algorithm found in MINPACK, plus support for bounds. R package version 1.2-1. https://cran.r-project.org/web/packages/minpack.lm/index.html. Accessed 8 Feb 2022.
Enciso-Rodriguez, F., D. Douches, M. Lopez-Cruz, J. J. Coombs, and G. de los Campos. 2018. Genomic selection for late blight and common scab resistance in tetraploid potato (Solanum tuberosum). G3 Genes|Genomes|Genetics 8:2471–2481.
Endelman, J.B., C.A. Schmitz Carley, P.C. Bethke, J.J. Coombs, M.E. Clough, W.L. da Silva, W.S. De Jong, D.S. Douches, C.M. Frederick, K.G. Haynes, D.G. Holm, J.C. Miller, P.R. Munoz, F.M. Navarro, R.G. Novy, J.P. Palta, G.A. Porter, K.T. Rak, V.R. Sathuvalli, A.L. Thompson, and G.C. Yencho. 2018. Genetic variance partitioning and genome-wide prediction with allele dosage information in autotetraploid potato. Genetics 209: 77–87.
Gehan, M.A., N. Fahlgren, A. Abbasi, J.C. Berry, S.T. Callen, L. Chavez, A.N. Doust, M.J. Feldman, K.B. Gilbert, J.G. Hodge, J.S. Hoyer, A. Lin, S. Liu, C. Lizarraga, A. Lorence, M. Miller, E. Planton, M. Tessman, and T. Sax. 2017. PlantCV v2: image analysis software for high-throughput plant phenotyping. PeerJ 5: e4088.
Gemenet, D.C., H. Lindqvist-Kreuze, B. De Boeck, G.S. Pereira, M. Mollinari, Z.B. Zeng, G.C. Yencho, and H. Campos. 2020. Sequencing depth and genotype quality: accuracy and breeding operation considerations for genomic selection applications in autopolyploid crops. Theoretical and Applied Genetics 133: 3345–3363.
Habyarimana, E., B. Parisi, and G. Mandolino. 2017. Genomic prediction for yields, processing and nutritional quality traits in cultivated potato (Solanum tuberosum L). Plant Breeding 136: 245–252.
International Commission on Illumination. 2019. Colorimetry – Part 4: CIE 1976 L*A*B* colour space. International Commission on Illumination.
Jones, C.R., T.E. Michaels, Schmitz Carley, C. Rosen, and L.M. Shannon. 2021. Nitrogen uptake and utilization in advanced fresh-market red potato breeding lines. Crop Science 61: 878–895.
Krupek, F.S., P.J. Dittmar, S.A. Sargent, L. Zotarelli, and D. Rowland. 2021. Impact of early potato desiccation method on crop growth, skinning injury, and storage quality maintenance. American Journal of Potato Research 98: 218–231.
Liu, Y., K. Lin-Wang, R. V. Espley, L. Wang, Y. Li, Z. Liu, P. Zhou, L. Zeng, and X. Zhang, Zhang, J., Allan, A.C. 2019. StMYB44 negatively regulates anthocyanin biosynthesis at high temperatures in tuber flesh of potato. Journal of Experimental Botany 70:3809–3824.
Menesatti, P., C. Angelini, F. Pallottino, F. Antonucci, J. Aguzzi, and C. Costa. 2012. RGB color calibration for quantitative image analysis: the “3D thin-plate Spline” warping approach. Sensors (Basel, Switzerland) 12: 7063–7079.
Moreda, G. P., M. A. Muñoz, M. Ruiz-Altisent, and A. Perdigones. 2012. Shape determination of Horticultural produce using two-dimensional computer vision – a review. Journal of Food Engineering 108:245–261.
Neilson, J.A.D., A.M. Smith, L. Mesina, R. Vivian, S. Smienk, and D. De Koyer. 2021. Potato tuber shape phenotyping using RGB imaging. Agronomy 11: 1781.
Parker, S.R., M.W. Shaw, and D.J. Royle. 1995. The reliability of visual estimates of disease severity on cereal leaves. Plant Pathology 44: 856–864.
Patel, K.K., A. Kar, and M.A. Khan. 2020. Development and an application of computer vision system for nondestructive physical characterization of mangoes. Agricultural Research 9: 109–124.
Pau, G., F. Fuchs, O. Sklyar, M. Boutros, and W. Huber. 2010. EBImage - an R package for image processing with applications to cellular phenotypes. Bioinformatics 26: 979–981.
Poland, J.A., and R.J. Nelson. 2011. In the eye of the beholder: the effect of rater variability and different rating scales on QTL mapping. Phytopathology 101: 290–298.
Prashar, A., C. Hornyik, V. Young, K. McLean, S.K. Sharma, M.F.B. Dale, and G.J. Bryan. 2014. Construction of a dense SNP map of a highly heterozygous diploid potato population and QTL analysis of tuber shape and eye depth. Theoretical and Applied Genetics 127: 2159–2171.
R Core Team. 2018. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria. https://www.R-project.org/. Accessed 8 Feb 2022.
Reeves, A.F. 1988. Varietal differences in potato tuber greening. American Potato Journal 65: 651–658.
Roe, M.R., J.L. Carlson, T.M. McManimon, A.D. Hegeman, and C. Tong. 2014. Differential accumulation and degradation of anthocyanins in Red Norland periderm is dependent on soil type and tuber storage duration. American Journal of Potato Research 91: 696–705.
Schlager, S. 2017. Morpho and rvcg - shape analysis in R. In Statistical shape and deformation analysis, eds. G. Zheng, S. Li, and G. Szekely., 217–256. Cambridge: Academic.
Schneider, C.A., W.S. Rasband, and K.W. Eliceiri. 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9: 671–675.
Selga, C., A. Koc, A. Chawade, and R. Ortiz. 2021. A bioinformatics pipeline to identify a subset of SNPs for genomics-assisted potato breeding. Plants 10: 30.
Si, Y., S. Sankaran, N.R. Knowles, and M.J. Pavek. 2017. Potato tuber length-width ratio assessment using image analysis. American Journal of Potato Research 94: 88–93.
Slater, A.T., G.M. Wilson, N.O. Cogan, J.W. Forster, and B.J. Hayes. 2014. Improving the analysis of low heritability complex traits for enhanced genetic gain in potato. Theoretical and Applied Genetics 127: 809–820.
Sood, S., Z. Lin, B. Caruana, A.T. Slater, and H.D. Daetwyler. 2020. Making the most of all data: combining non-genotyped and genotyped potato individuals with HBLUP. The Plant Genome 13: e20056.
Stefaniak, T.R., S. Fitzcollins, R. Figueroa, A.L. Thompson, Schmitz Carley, and L.M. Shannon. 2021. Genotype and variable nitrogen effects on tuber yield and quality for red fresh market potatoes in Minnesota. Agronomy 11: 255.
Stich, B., and D. Van Inghelandt. 2018. Prospects and potential uses of genomic prediction of key performance traits in tetraploid potato. Frontiers in Plant Science 9: 159. https://doi.org/10.3389/fpls.2018.00159.
Sverrisdóttir, E., S. Byrne, E.H.R. Sundmark, H.O. Johnsen, H.G. Kirk, T. Asp, L. Janss, and K.L. Nielsen. 2017. Genomic prediction of starch content and chipping quality in tetraploid potato using genotyping-by-sequencing. Theoretical and Applied Genetics 130: 2091–2108.
Sverrisdóttir, E., E.H.R. Sundmark, H.Ø. Johnsen, H.G. Kirk, T. Asp, L. Janss, G. Bryan, and K.L. Nielsen. 2018. The value of expanding the training population to improve genomic selection models in tetraploid potato. Frontiers in Plant Science 9: 1118. https://doi.org/10.3389/fpls.2018.01118.
USDA Plant Variety Protection Office. 2015. USDA form OMB NO 0581 – 0055. USDA-AMS. https://www.ams.usda.gov/resources/st470-potato. Accessed 8 Feb 2022.
Van der Werff, H.M.A., and F.D. Van der Meer. 2008. Shape-based classification of spectrally identical objects. ISPRS Journal of Photogrammetry and Remote Sensing 63: 251–258.
Van Eck, H.J., J.M. Jacobs, P. Stam, J. Ton, W.J. Stiekema, and E. Jacobsen. 1994. Multiple alleles for tuber shape in diploid potato detected by qualitative and quantitative genetic analysis using RFLPs. Genetics 137: 303–309.
Zhou, L., V. Chalana, and Y. Kim. 1998. PC-based machine vision system for real‐time computer‐aided potato inspection. International Journal of Imaging Systems and Technology 9: 423–433.
Acknowledgements
We are very grateful to Maria Caraza Harter who helped us figure out how to set up a lightbox. We would like to thank the many members of the Shannon lab who helped with grading, visual rating, and taking images including: Husain Agha, Katelyn Filbrandt, Sophia Fitzcollins, Jessica Huege, Colin Jones, John Larsen, Thomas McGehee, Xiaoxi Meng, Laura Schulz, Thomas Stefaniak, Heather Tuttle, and Muyideen Yusuf. Keith Mann managed our field site at Grand Rapids MN, and Ron Faber managed our field site in Becker MN. Finally, four anonymous reviewers and the editor provided insightful feedback on earlier drafts of the manuscript.
Funding
This work was funded by USDA-ARS 58-3060-0-012 AMD 1, USDA-NIFA 2016-34141-25707, USDA-NIFA 2019-34141-30284, and the Minnesota Department of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Dr. Cari Schmitz Carley works for Aardevo a diploid potato breeding company. The views expressed are those of the authors and do not necessarily reflect the position or policy of Aardevo.
Dr. Max Feldman and Dr. Darrin Haagenson work for the USDA-ARS. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA-ARS is an equal opportunity provider and employer.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
ESM 1
(PDF 3.70 MB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Miller, M.D., Schmitz Carley, C.A., Figueroa, R.A. et al. TubAR: an R Package for Quantifying Tuber Shape and Skin Traits from Images. Am. J. Potato Res. 100, 52–62 (2023). https://doi.org/10.1007/s12230-022-09894-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12230-022-09894-z