Abstract
In this study, a unicellular soil alga isolated from farmland in Germany was surveyed. The investigation of the hypervariable molecular markers ITS1 rDNA and ITS2 rDNA identified strain E71.10 as conspecific with Vischeria sp. SAG 51.91 (Eustigmatophyceae). The culture was tested for biomass generation and for the yield of fatty acids and amino acids. The survey included four different culture conditions (conventional, elevated CO2, nitrogen depletion, or sodium chloride stress) at room temperature. The best yield of dry biomass was achieved applying 1% CO2, whereas nitrogen-free medium resulted into least growth. The fatty acid content peaked in nitrogen-free medium at 59% per dry mass. Eicosapentaenoic acid was the most abundant fatty acid in all treatments (except for nitrogen free), accounting for 10.44 to 16.72 g/100 g dry mass. The highest content of amino acids (20%) was achieved under conventional conditions. The results show that abiotic factors strongly influence to which extent metabolites are intracellularly stored and they confirm also for this yet undescribed strain of Vischeria that Eustigmatophyceae are promising candidates for biotechnology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Eustigmatophyceae have experienced increasing attention in the last years, because they turned out to be biotechnologically promising in terms of biomass generation, production of lipids, pigments, and other compounds (Stoyneva-Gärtner et al. 2019). Li et al. (2012) screened several species and found high levels of β-carotene (up to 5.9% dry mass) and biomass production rates up to 9.8 g/L in bubble column photobioreactors. Gao et al. (2016) reported a lipid production of 0.28 g/L/day for Vischeria stellata and stressed that this alga can be a source for nutraceuticals or used for biodiesel production. Moreover, a large proportion of the oleaginous components are polyunsaturated fatty acids, and significant amounts of the valuable eicosapentaenoic acid (EPA) have been reported for some of these algae (Cepák et al. 2014; Wang et al. 2018).
In this study, a yet undescribed strain of a eustigmatophycean alga isolated from field soil was tested for its potential of fatty and amino acid production. Both compound classes are of biotechnological interest. The taxonomic position was evaluated by using four molecular markers (18S rDNA, rbcL, ITS1 rDNA, ITS2 rDNA) which placed this alga into the genus Vischeria. Furthermore, lipid production was optimized during cultivation by applying stress protocols such as nitrogen depletion and addition of sodium chloride. To our knowledge, this alga has never been analyzed biochemically and phylogenetically before. Here, we showed that the strain Vischeria sp. E71.10 exhibited high growth rates and abundantly accumulated fatty acids, particularly eicosapentaenoic acid.
Material and methods
A eustigmatophycean microalga was isolated by W. Oesterreicher (Kitzbühel, Austria) from field soil close to Ahlum (Wolfenbüttel, Lower Saxony, Germany) in course of a study testing changes of soil algae biocoenosis depending on the extent of insecticides and fertilization (Sautthof and Oesterreicher 1994). The strain was deposited as E71.10 at the algae culture collection at the Institute of Botany, University of Innsbruck, however was not listed in the strain catalog (ASIB; Gärtner 1996). The cell morphology was observed by light microscopy (Nikon Eclipse 80i, objective Plan Apo VC 100 × 1.40, camera DS 5 M; Nikon Instruments, Amsterdam, Netherlands) using either differential interference contrast or fluorescence mode (filter em = 600 LP, ex = 480/40). Prior use, the strain was grown at 15 °C and approximately 40 to 50 μmol photons/m2/s (14 h light, 10 h darkness) provided by full spectrum fluorescence 18 W tubes (Narva BioVital 958, Plauen, Germany). Modified Triple Nitrate Bolt’s Basal Medium (‘3 N BBM’) was used according to the recipe of the CCCryo strain collection (Potsdam, Germany, http://cccryo.fraunhofer.de/sources/files/medien/BBM.pdf). For the generation of biomass, the cells were cultivated at room temperature in 1-L glass column bubble reactors aerated with compressed air at 0.6 mL/min. The fluorescence tube illumination (14 h/day) was approximately 220 μmol photons/m2/s. Growth was evaluated by measuring the absorbance of a 3-mL subsample at 750 nm with a spectrophotometer (Lange Xion 500, Germany). The pH was monitored with a WTW electrode (Xylem Instruments, Weilheim, Germany). In alternative setup, the Erlenmeyer flasks with 3 N BBM medium were put into a growth chamber enriched with constant 1% CO2 supply (Percival SE-41AR3, CLF PlantClimatics, Germany). Two stress treatments for enhancement of fatty acid content were performed either by a complete medium change to nitrogen-free BBM (‘-N BBM’) or by adding 4 g/L NaCl to 3 N BBM. For harvesting, the reactors were discharged and the cell concentrated by centrifugation (3000g, 15 min). The algal pellet was immediately frozen at − 80 °C and subsequently lyophilized at darkness for 48 h.
Analytics
The fatty acid (FA) content was measured by gas chromatography (GC) using a modified transmethylation protocol of Welz et al. (1990): 5 mg of lyophilized algae powder was suspended and methylated with 5 mL of methanol/acetyl chloride (p.a. grade) with a volumetric ratio of 50:1 for 4 h at 60 °C. The reaction was stopped by slowly adding 2.5 mL of a potassium carbonate solution (p.a. grade, 60 g/L). The resulting fatty acid methyl esters were extracted by adding 2 mL of hexane (GC/MS grade) and shaking for 2 min. After phase separation, 1 mL of the supernatant phase, containing the methyl esters, was transferred in a 1.5-mL crimp vial and stored at − 18 °C until measurement. The hexane-extract was injected in a Thermo Trace 1300 GC (Thermo Scientific), equipped with an autosampler AS 1310 and an SSL injector and flame ionization detector (FID). The chromatographic conditions were as follows: injection volume 1 μL, injector temperature was set at 240 °C. Helium was used as carrier gas (constant flow) with 1.5 mL/min and a split flow at 30 mL/min. An Agilent J&W capillary column DB-23 60 m, 0.25 mm ID, and 0.25 μm film thickness were used for analytical separation. The oven temperature gradient was 0–3 min 130 °C; 6.5 °C/min to 170 °C. 2.8 °C/min to 214 °C and held for 12 min. Then, 3 °C/min to 240 °C and held for 15 min. The FID was set at a temperature of 280 °C, 450 mL/min air flow, 45 mL/min hydrogen flow, and nitrogen as make up gas at 40 mL/min. Data analysis was performed with Chromeleon V7.2 (Thermo Scientific). The calibrations were done with an external standard (Supelco Fame Mix C14 - C22, Eicosapentaenoic acid methyl ester, Sigma-Aldrich). Determination of amino acid (AA) profiles. For protein digestion, a modified acidic hydrolyzation was performed according to Fountoulakis and Lahm (1998): 10 mg lyophilized algae sample was hydrolyzed with 1 mL of hydrochloric acid (c = 6 mol/L, analytical grade) in a 1.5-mL tight screw cap glass vial for 24 h at 110 °C. The hydrolysate was diluted appropriately in deionized water and clarified by centrifugation at 25,000g for 10 min. The clear supernatant was transferred in 2-mL HPLC vials using a final dilution in a range from 20-fold to 60-fold and stored at 4 °C prior analysis. The amino acids were measured on an Agilent 1260 Infinity II HPLC preforming an OPA-3-MPA (o-phthaldialdehyde 3-mercaptopropionic acid) and FMOC-CL (fluorenylmethyloxycarbonyl chloride) pre-column derivatization and subsequent fluorescence detection (Schuster 1988). The stationary phase was a Thermo ODS Hypersil 2 column 4.6 × 250 mm kept at 40 °C. The injection volume was 10 μL and the pump flow rate was set to 1.5 mL/min and consisted of mobile phases (A) sodium dihydrogen phosphate buffer (c = 0.04 mol/L, analytical grade) and (B) acetonitrile, methanol (both in analytical grade) and deionized water in volumetric ratio of 45:45:10. The applied linear gradient was 0–3 min 2% B, 18 min 72% B, 18.1–25 min 100% B, re-equilibration time 6 min. For the two-step column derivatization of the sample, the following three reagents have been applied: (1) borate-buffer: made of ortho-boric acid, c = 0.4 mol/L at pH 10.2, adjusted with aqueous potassium hydroxide solution (c = 10 mol/L), syringe filtered (0.45 μm polyamide); (2) OPA-3-MPA reagent (Sigma-Aldrich): from 10 mg OPA in 1-mL borate buffer adding 10 μL 3-MPA; and (3) FMOC-CL reagent (Merck, Darmstadt, Germany): 2.5 mg in 1 mL acetonitrile. All chemicals were analytical standard grade. The final solutions were stored at 4 °C and useable for 1 week. Prior to the column injection, the injector program according to Table 1 was applied in the sample vial for adding the fluorophores, where FMOC derivatization was done for proline and OPA for the other measured amino acids.
The detection of amino acids was performed by HPLC using fluorescence detection at ex/em 340/450 nm for OPA-3-MPA and ex/em 266/305 nm for FMOC-CL, the later especially applied for proline. The calibration for amino acid quantification was performed by external standardization with the amino acid standard mixture AAS18 and further single analytical standards from Merck. Asparagine and glutamine were hydrolyzed into their acids and are thus not detectable. Data acquisition and analysis were performed with OpenLAB CDS ChemStation Edition software (Agilent Technologies). All analytical samples were determined in biological and analytical triplets. Mean values were calculated by the arithmetic mean, standard deviations were calculated presuming a normal distribution and resulting the coefficient of variation was shown. The yield was calculated as compound production rate per day of reactor life time.
Molecular methods
Total genomic DNA of the strain E71.10 was extracted according to Procházková et al. (2018). The 18S small subunit ribosomal RNA gene (18S rDNA), internal transcribed spacer regions 1 and 2 (ITS1 rDNA, ITS2 rDNA), and ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene regions were amplified from DNA isolates by polymerase chain reaction using existing primers (Table S1). Amplification and sequencing reactions for these markers were identical to those described by Procházková et al. (2018). Nuclear rDNA regions of ITS2 were identified using the web interface for hidden Markov model-based annotation (Keller et al. 2009) at the ITS2 database (Ankenbrand et al. 2015; http://its2.bioapps.biozentrum.uni-wuerzburg.de/). The sequence was then folded with 5.8S–LSU stem regions using the Mfold server accessible at http://mfold.rna.albany.edu/?q5mfold (Zuker 2003). A model of the secondary structure consistent with the specific features of nuclear rDNA ITS2 was selected: four helixes and U–U mismatch in helix II (Coleman 2007). For detecting compensatory base changes (CBCs), the ITS2 sequences were aligned based on a sequence-structure analysis (Schultz and Wolf 2009) using 4SALE (Seibel et al. 2006, 2008). The secondary structure of nuclear rDNA ITS2 was drawn using VARNA version 3.9 (Darty et al. 2009). The correlation between the CBC criterion and the biological species concept was introduced by Coleman (2000) and was statistically proven by Müller et al. (2007): if a CBC is found between two organisms classified within the same genus, then they are two different species with 93% probability. Presence of CBC was checked between the strain E71.10 and its closest relative strain SAG 51.91. The obtained sequences were submitted to the National Center for Biotechnology Information (NCBI) Nucleotide sequence database (accession numbers 18S rDNA: MN781105; ITS1 rDNA + 5.8S rDNA + ITS2 rDNA: MN781106; rbcL: MN781107).
Results
Morphological and molecular characterization
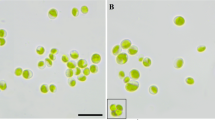
Young cells of strain E71.10 were solitary elongate and more or less characteristically bean shaped (Fig. 1a); they became larger and changed to irregularly ellipsoidal to almost spherical appearance (Fig. 1b). Wall surfaces were smooth; bulges typically for some Eustigmatophyceae occurred rarely. At old stages, the alga turned from green to orange showing a prominent extraplastidal pyrenoid. A bi- or tri-layered cell wall was visible (Fig. 1c). Most of the cells were immotile in both liquid cultures and on agar, but flagellates occurred occasionally (Supplementary Video 1). The latter had two visible flagella, one of them very short. All cells had a single parietal chloroplast, cup-shaped with incisions or lobes; however, no eyespot was detected. Cell division by autosporangia was observed.
The 18S rDNA sequence of strain E71.10 was 100% identical with several strains assigned to three different genera Eustigmatos, Vischeria, and Chloridella (Supplementary Table S2). One of them was Vischeria sp. CAUP Q 202 (Yurchenko et al. 2016). For rbcL, strain E71.10 was closely related to Vischeria helvetica UTEX 49 (Yang et al. 2012). Moreover, the alga was almost identical for the hypervariable markers ITS1 and ITS2 rDNA with Vischeria sp. SAG 51.91, isolated from snow detritus in the Belianské Tatry Mountains (Slovakia) (Supplementary Table 2). ITS2 rRNA secondary structure of strain E71.10 and SAG 51.91 was compared; no CBC was found in the entire structure (Supplementary Fig. 1).
Biomass, fatty, and amino acid yield
Table 2 shows the total harvested dry mass (DM) of Vischeria sp. E71.10, the daily dry mass production rate, and the total content of fatty acids, respectively, amino acids per dry mass for all four culture treatments. The by far best yield of dry biomass was achieved applying elevated CO2 (1%) in the atmosphere of the chamber in combination with 3 N BBM medium, whereas nitrogen-free medium resulted into least growth. The fatty acid content of approx. 42% for 3NBBM was outraged when exposing cells to nitrogen-free medium (-NBBM) resulting into almost 59% FAs per dry mass. On the other hand, -NBBM showed the least growth and biomass accumulation rates, succeeded by sodium chloride stress as the second least. The latter hardly changed the relative cellular FA and amino acid content compared to the standard treatment.
By means of GC, the abundance of fatty acids was compared among the four treatments (Table 3). The sizes of main fatty acids ranged from C14:0 to C20:4, and they were qualitatively similar between all treatment except the nitrogen-free exposure; in the latter case, C16:1 (palmitoleic acid) was particularly increased. In all other assays, C20:5 (eicosapentaenoic acid, EPA) was the most abundant fatty acid in a range of 10.4 to 16.7% of dry mass.
The spectrum of cellular amino acids was measured by HPLC as shown in Table 4. A broad range of AAs was found, and the sodium chloride stress did hardly change the absolute level of about 20% per dry mass. However, during the fast growth under CO2 application, AA content decreased to approx. 10% and was even less (5.5%) during nitrogen depletion.
Discussion
Cell morphology and taxonomy
In Eustigmatophyceae, the 18S rDNA marker has a resolution useful for definition of the new clades (e.g., Goniochloridales) but is not suitable for species determination (Fawley et al. 2014). Instead, the use of a polyphasic approach including the more variable ITS2 rDNA marker is more reliable for defining species boundaries in microalgae (Kryvenda et al. 2018). The investigated strain E71.10 was almost identical for hypervariable markers ITS1 and ITS2 with strain SAG 51.91. At the LM level, strain E71.10 shared with SAG 51.91 vegetative cells without wall projections, bean shaped to spherical during cell aging, one single plastid per cell throughout the cell cycle; however, for the latter, the formation of zoospores was not observed (Kryvenda et al. 2018). In Eustigmatos and Vischeria, the flagellated stages possess a single emergent flagellum. In strain E71.10, two visible flagella were present, one of them very short. Using the key of Ettl and Gärtner (2014), the ellipsoidal to spherical smooth cell shape in E71.10 cells resembled Pseudellipsoidion (Neustupa and Němcová 2001).
In the 18S rDNA phylogeny of Eustigmatophyceae, strains previously assigned to Eustigmatos, Vischeria, and Chloridella were recovered within a well-supported monophyletic clade, the ʿEustigmataceae Groupʾ sensu (Fawley et al. (2014). Based on the sequence-structure ITS2 rDNA phylogeny, this latter group was further distinguished into a ʿVischeria cladeʾ and a ʿVischeria-related cladeʾ (Kryvenda et al. 2018). Consequently, all taxa of the ʿVischeria cladeʾ should be transferred into the genus Vischeria (Kryvenda et al. 2018). Therefore, strain E71.10 of this study was placed to Vischeria, but no species assignation was possible and no species description was attempted in this study. A better resolution for the genetic distinction of species within Vischeria clade was achieved using ITS2 rDNA marker. However, the presence of multiple paralogous sequences within one strain, i.e., an intragenomic ITS2 sequence variation, was revealed for Eustigmatophyceae (Kryvenda et al. 2018), which makes further taxonomic assignments among investigated Vischeria strains impossible, until the divergence of intragenomic ITS2 paralogues in this genus is better understood.
Biomass, fatty, and amino acid yield
The faster growth and productivity rates were achieved with 3 N BBM and elevated amounts of CO2, at almost 0.2 g per liter reactor volume per day, which is in accordance with many other studies (e.g., Wang et al. 2018). To increase the yield of fatty acids, the logarithmic growth phase should be followed by exposure to sodium chloride and nitrogen depletion before harvest. Wang et al. (2018) found high amount of palmitoleic acid (C16:1) in several Eustigmatophyceae, ranging from 15% DM to approx. 25% DM under decreasing nitrogen availability. Here, this FA was the second most abundant, respectively, most dominant FA in case of -N BBM, and with comparable amounts to the reference. However, the yield was lower when applying 3 N BBM without additional CO2 and worst under sodium chloride stress reaching only about 5% DW. Remarkable are the high contents of the valuable EPA; however, its concentration was not elevated by application of nitrogen stress. Except for the treatment with -NBBM, the EPA content was higher (expressed as % in total fatty acids) by 10 to 20% when compared to for ITS2 rDNA almost genetically identical strain Vischeria sp. SAG 51.91 (Lang et al. 2011). In the eustigmatophycean alga Trachydiscus guangdongensis, EPA was also the most prominent FA, but constantly decreased during cultivation (Gao et al. 2019). In a study of Cepák et al. (2014) with the same species, EPA content was highest at the lowest light level they applied (100 μmol photosynthetic active radiation (PAR)/m2/s). In Vischeria sp. E71.10, EPA was produced fastest in high nitrogen medium and with additional application of CO2. The production rate of 32.32 mg/L/day was extraordinarily high compared to other studies involving Eustigmatophyceae. Qunju et al. (2016) reported only a maximum of 9.64 mg/L/day for total poly unsaturated fatty acid yield of a high productivity strain, Nannochloropsis sp.
The applied HPLC method proofed to be both economic and feasible for the determination of algal amino acids. Chua and Schenk (2017) reviewed methods to economically harvest proteins from Eustigmatophyceae used as an alternative source for animal feed and foodstuff. Generally, studies about individual AA spectra are sparse. Tibbetts et al. (2015) is one of the few studies analyzing a Eustigmatophyceae, Nannochloropsis granulata, and like in this study, glutamic acid was the most abundant AA.
Conclusions
The molecular ITS2 rDNA marker was useful for placing strain into E71.10 to Vischeria. The culture performed well in terms of high growth rates and abundant fatty acid accumulation, namely EPA, confirming the biotechnological potential and oleaginous character of such eustigmatophycean microalgae. Moreover, old cultures turning from green to orange indicate carotenoid accumulation during nitrogen starvation. These facts underline the potential of this strain as a candidate for sustainable bioresources. Further investigations should include an optimization of the reactor settings for enhanced yield and testing the pigment (carotenoid) production as an additional valuable biocompound.
References
Ankenbrand MJ, Keller A, Wolf M, Schultz J, Förster F (2015) ITS2 database V: twice as much. Mol Biol Evol 32:3030–3032. https://doi.org/10.1093/molbev/msv174
Cepák V, Přibyl P, Kohoutková J, Kaštánek P (2014) Optimization of cultivation conditions for fatty acid composition and EPA production in the eustigmatophycean microalga Trachydiscus minutus. J Appl Phycol 26:181–190. https://doi.org/10.1007/s10811-013-0119-z
Chua ET, Schenk PM (2017) A biorefinery for Nannochloropsis: induction, harvesting, and extraction of EPA-rich oil and high-value protein. Bioresour Technol 244:1416–1424. https://doi.org/10.1016/j.biortech.2017.05.124
Coleman AW (2000) The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist 151:1–9. https://doi.org/10.1078/1434-4610-00002
Coleman AW (2007) Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res 35:3322–3329. https://doi.org/10.1093/nar/gkm233
Darty K, Denise A, Ponty Y (2009) Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25:1974–1975. https://doi.org/10.1093/bioinformatics/btp250
Ettl H, Gärtner G (2014) Syllabus der Boden-, Luft- und Flechtenalgen. Springer, Berlin. https://doi.org/10.1007/978-3-642-39462-1
Fawley KP, Eliáš M, Fawley MW (2014) The diversity and phylogeny of the commercially important algal class Eustigmatophyceae, including the new clade Goniochloridales. J Appl Phycol 26:1773–1782. https://doi.org/10.1007/s10811-013-0216-z
Fountoulakis M, Lahm HW (1998) Hydrolysis and amino acid composition analysis of proteins. J Chromatogr A 826:109–134. https://doi.org/10.1016/S0021-9673(98)00721-3
Gao B, Yang J, Lei X, Xia S, Li A, Zhang C (2016) Characterization of cell structural change, growth, lipid accumulation, and pigment profile of a novel oleaginous microalga, Vischeria stellata (Eustigmatophyceae), cultured with different initial nitrate supplies. J Appl Phycol 28:821–830. https://doi.org/10.1007/s10811-015-0626-1
Gao B, Huang L, Wang F, Zhang C (2019) Trachydiscus guangdongensis sp. nov., a new member of Eustigmatophyceae (Stramenopiles) isolated from China: morphology, phylogeny, fatty acid profile, pigment, and cell wall composition. Hydrobiologia 835:37–47. https://doi.org/10.1007/s10750-019-3925-8
Gärtner G (1996) ASIB – the culture collection of algae at the botanical Institute of the University at Innsbruck (Austria). − catalogue of strains 1996. Ber nat-med Verein Innsbruck 83:45–69
Keller A, Schleicher T, Schultz J, Müller T, Dandekar T, Wolf M (2009) 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene 430:50–57. https://doi.org/10.1016/j.gene.2008.10.012
Kryvenda A, Rybalka N, Wolf M, Friedl T (2018) Species distinctions among closely related strains of Eustigmatophyceae (Stramenopiles) emphasizing ITS2 sequence-structure data: Eustigmatos and Vischeria. Eur J Phycol 53:471–491. https://doi.org/10.1080/09670262.2018.1475015
Lang I, Hodač L, Friedl T, Feussner I (2011) Fatty acid profiles and their distribution patterns in microalgae: a comprehensive analysis of more than 2000 strains from the SAG culture collection. BMC Plant Biol 11:124. https://doi.org/10.1186/1471-2229-11-124
Li Z, Sun M, Li Q, Li A, Zhang C (2012) Profiling of carotenoids in six microalgae (Eustigmatophyceae) and assessment of their ß-carotene productions in bubble column photobioreactor. Biotechnol Lett 34:2049–2053. https://doi.org/10.1007/s10529-012-0996-2
Müller T, Philippi N, Dandekar T, Schultz J, Wolf M (2007) Distinguishing species. RNA 13:1469–1472 http://www.rnajournal.org/cgi/doi/10.1261/rna.617107
Neustupa J, Němcová Y (2001) Morphological and taxonomic study of three terrestrial eustigmatophycean species. Beih Nova Hedwigia 123:373–386
Procházková L, Remias D, Řezanka T, Nedbalová L (2018) Chloromonas nivalis subsp. tatrae, subsp. nov. (Chlamydomonadales, Chlorophyta): re-examination of a snow alga from the high Tatra Mountains (Slovakia). Fottea 18:1–18. https://doi.org/10.5507/fot.2017.010
Qunju H, Wenzhou X, Fangfang Y, Tao L, Guanghua W, Shikun D, Hualian W, Jiewei F (2016) Evaluation of five Nannochloropsis sp. strains for biodiesel and poly-unsaturated fatty acids (PUFAs) production. Curr Synthetic Sys Biol 4:128. https://doi.org/10.4172/2332-0737.1000128
Sautthof W, Oesterreicher W (1994) Untersuchungen über den Einfluß einer intensive Pflanzenproduktion auf die Zusammensetzung der Bodenalgenflora. In: Bartels G, Kampmann T (eds) Effects of a long-term application of plant protection products used in different intensities and development of assessment criterions. Blackwell-Wiss.-Verlag, Berlin/Wien, pp 167–191
Schultz J, Wolf M (2009) ITS2 sequence-structure analysis in phylogenetics: a how-to manual for molecular systematics. Mol Phylogenet Evol 52:520–523. https://doi.org/10.1016/j.ympev.2009.01.008
Schuster R (1988) Determination of amino acids in biological, pharmaceutical, plant and food samples by automated precolumn derivatization and high-performance liquid chromatography. J Chromatogr B 431:271–284. https://doi.org/10.1016/s0378-4347(00)83096-0
Seibel PN, Müller T, Dandekar T, Schultz J, Wolf M (2006) 4SALE--a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinformatics 7:498. https://doi.org/10.1186/1471-2105-7-498
Seibel PN, Müller T, Dandekar T, Wolf M (2008) Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE. BMC Res Notes 1:91. https://doi.org/10.1186/1756-0500-1-91
Stoyneva-Gärtner M, Uzunov B, Gärtner G, Borisova C, Draganova P, Radkova M, Stoykova P, Atanassov I (2019) Current bioeconomical interest in stramenopilic Eustigmatophyceae: a review. Biotechnol Biotec Equ 33:302–314. https://doi.org/10.1080/13102818.2019.1573154
Tibbetts SM, Milley JE, Lall SP (2015) Chemical composition and nutritional properties of freshwater and marine microalgal biomass cultured in photobioreactors. J Appl Phycol 27:1109–1119. https://doi.org/10.1007/s10811-014-0428-x
Wang F, Gao B, Huang L, Su M, Dai C, Zhang C (2018) Evaluation of oleaginous eustigmatophycean microalgae as potential biorefinery feedstock for the production of palmitoleic acid and biodiesel. Bioresour Technol 270:30–37. https://doi.org/10.1016/j.biortech.2018.09.016
Welz W, Sattler W, Leis HJ, Malle E (1990) Rapid analysis of non-esterified fatty acids as methyl esters from different biological specimens by gas chromatography after one-step esterification. J Chromatogr B 526:319–329. https://doi.org/10.1016/S0378-4347(00)82516-5
Yang EC, Boo GH, Kim HJ, Cho SM, Boo SM, Andersen RA, Yoon HS (2012) Supermatrix data highlight the phylogenetic relationships of photosynthetic stramenopiles. Protist 163:217–231. https://doi.org/10.1016/j.protis.2011.08.001
Yurchenko T, Ševčíková T, Strnad H, Butenko A, Eliáš M (2016) The plastid genome of some eustigmatophyte algae harbours a bacteria-derived six-gene cluster for biosynthesis of a novel secondary metabolite. Open Biol 6:160249. https://doi.org/10.1098/rsob.160249
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. https://doi.org/10.1093/nar/gkg595
Acknowledgments
Open access funding provided by University of Applied Sciences Upper Austria. We thank Dr. C. Griesbeck, Management Center Innsbruck (MCI), Austria, for providing the strain. We thank Marek Eliáš (University of Ostrava, Czech Republic) for providing sequences of the two primers (ITS-F-Visch, ITS4k-Eustig).
Funding
This research was funded by the Austrian Science Fund (FWF) project P29959 granted to D.R.
Author information
Authors and Affiliations
Contributions
Study design: KK, BM; performed research: BM, CN, DR, LP; analyzed data: CN, KK, LP; paper writing: DR, LP, KK, CN, LN
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Remias, D., Nicoletti, C., Krennhuber, K. et al. Growth, fatty, and amino acid profiles of the soil alga Vischeria sp. E71.10 (Eustigmatophyceae) under different cultivation conditions. Folia Microbiol 65, 1017–1023 (2020). https://doi.org/10.1007/s12223-020-00810-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-020-00810-8