Abstract
The biodiversity protection and monitorning is one of main goals of natural history musems worldwide. Conservation issues are particularly important for freshwater fish which are one of the most threatened taxa for the consequences of climate change and human activies. In Mozambique freshwater rivers are poorly explored and the impact of aquaculture and human activities on local biodiversity in almost unknown. Here we propose the barcoding analysis of cytochrome c oxidase I (COI) mitochrondrial DNA of 41 frehswater fishes catched in four rivers of southern Mozambique and 53 from a museum collection. As evidence of previous knowledge gaps, barcoding results revealed twenty new haplotypes described for the first time in the taxa Cichlidae and Amphilidae. From a methodological point of view, the barcoding approach demonstrated a critical point connected to the requested 650 bp length of amplified sequences. In fact, high weight genomic DNA is unattainable from museum samples and also in wildlife samples collected in pristine rivers. For this reason we furtherly tested the efficiency of DNA mini-barcoding analysis for 53 fish from a museum collection. The Mini-barcode method retrieved 56.6% of sequences successfully analyzed versus 3% of barcoding. The high performance of this thecniques is discussed in relation to biodiversity monitoring and to fill the taxonomy gaps in museum collections.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Molecular approaches are innovative tools helpful to identify new taxa of the fish fauna. In particular, molecular techniques based on DNA barcoding can be used to study taxonomy and resolve evolutionary relationships among taxa (Waugh 2007) to define a correct species identification and origin. DNA barcoding techniques are nowadays widely applied in different fields as product labelling accuracy in commercial fish and seafood (Filonzi et al. 2010, 2021), identification of threatened animal species used in wildlife criminal trade (Staats et al. 2016) and to discover the biodiversity level between and within different animal taxa (Bezeng and Bank 2019). This approach is also valuable to identify species displaying low interspecific divergence, such as the ones belonging to the megadiverse neotropical freshwater fish fauna (Pereira et al. 2013).
The application of genetic studies for the identification of African freshwater fish is an urgent need considering the potential richness of species and the paucity of available data in the field of taxonomy, population and conservation genetics (Pereira et al. 2013). Focusing the attention on specific African countries, most of genetic studies have examined aquaculture stocks or introduced populations instead of studying the natural ones (Mojekwu et al. 2021). That is the case of Mozambique, where freshwater fish biodiversity has been rarely investigated in the past and many gaps are still waiting to be filled out. For this reason, in this work we selected a dual approach investigating two different fish taxa of Mozambique dealing with issues either in the commercial field or in natural sciences: respectively species belonging to the Cichlidae and the Amphilidae families.
Among the Cichlidae family, Tilapia is one of the most complex genera with three subgenera taxonomically distinguished mainly by reproductive and feeding behavior: Oreochromis (female mouthbrooders), Tilapia (biparental and substrate spawners), and Sarotherodon (biparental mouthbrooders) (Yáñez et al. 2020). The evolution history of these taxa is still unresolved. In fact, a taxonomic approach based on behaviour and ecological habits certainly underestimates the complexity and genetic diversity within the taxon. Despite this, genetic manipulation and commercial spreading of tilapiines is increasing (Yáñez et al. 2020). In fact, they are becoming invasive alien species worldwide and knowing their genetic profiles is becoming fundamental (Cassemiro et al. 2018).
In Mozambique, five species of Tilapia are nowadays described, namely Oreochromis mossambicus, Oreochromis aureous, Coptodon rendalli, Oreochromis mortimeri and Oreochromis placidus. O. mossambicus is the most cultivated and studied species worldwide, because of its ability to adapt to various habitats and environmental fluctuations in terms of salinity, temperature and dissolved oxygen (Russell et al. 2012). However, the lack of information and mapping of native species of cichlids in water systems of Mozambique generated several conservation issues also referred to the introduction of exotic tilapines into the natural environment, which may harm native endemic fish. In fact, aquaculture has been growing significantly in recent years in Mozambique, because of the government call to increase family income and to reduce pressure from fishing at sea.
At the same time, different river and stream systems remain poorly explored and deserve more attention to be dedicated to conservation issues. In fact, recent results obtained in several fish species from mountain streams in Southern Africa have shown high diversity at species level and taxonomic conflicts (Chakona et al. 2018; Schmidt et al. 2015,). For this reason, in this work further attention has been dedicated to the understudied genus Amphlilius, which is a cryptic species, one of the smallest of Siluriformes inhabiting a still circumscribed geographic range (Chakona et al. 2018). Four new species of Amphilius were recently described by Mazungula and Chakona (2021) and evidence of the existence of other genetic lineages of the Amphilidae family has been supposed. The opportunity of studying such species in pristine areas of Mozambique could also provide insights on the patterns of endemism of African stream fishes.
Based on the above considerations, the opportunity of studying samples from museum collections coupled to natural resources by means of a molecular genetic approach is presented. In particular, a molecular characterization of Cichlidae and Amphilidae families sampled in Southern Mozambique rivers and in the Natural History Museum of Maputo was carried out to define the specific taxonomy through a double approach in relation to the DNA quality. In fact, samples from museum collections and pristine natural sites often display degraded DNA (Whitfield 1999; Hajibabaei et al 2006). For this reason, both COI barcoding and mini-barcoding were applied to fish samples to fill knowledge gaps and complete the species identification (Sultana et al. 2018).
2 Material and methods
2.1 Sampling area
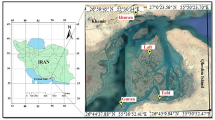
Forty-one freshwater fish samples were collected in 2018 and 2019 by traditional methods using nets and fishermen activities in four different rivers of Southern Mozambique (Fig. 1 and Table 1). In addition, 53 specimens belonging to the Natural History Museum (NHM) of Maputo were also obtained and analyzed.
2.2 Sampling and extraction
For each individual, a small fragment of caudal fin was clipped and stored in 95% ethanol. Genomic DNA was extracted and purified by Wizard® Genomic DNA Purification Kit (Promega) and DNA quality was visually inspected by 1% agarose gel electrophoresis in TAE buffer before amplification of Cytochrome Oxidase I (COI) mitochondrial gene.
2.3 PCR amplification and sequencing
A fragment of COI mtDNA gene was amplified using the standard primers FF2d and FR1d for the regular barcoding described by Ivanova et al. (2007). Polymerase chain reaction (PCR) amplification was performed following Filonzi et al. (2010) in a final volume of 25 μl consisting of 30 ng of genomic DNA, 1 U of Biotherm Red DNA Polymerase (Fisher Molecular Biology), Mg2 + 2.5 mM, 10 pmol of each primer, 1 × reaction buffer and dNTPs 0.2 mM. PCR cycles were conducted with an initial 5 min denaturation step at 94 °C, 35 cycles of 30 s at 94 °C, 45 s at 52 °C and 1 min at 72 °C, final extension at 72 °C for 10 min.
Alternatively, a shorter fragment of COI mtDNA was amplied using the mini-barcode universal primers for fish species described in Sultana et al. (2018): Fish_miniFW: ATCACAAAGACATTGGCACCCT and Fish_miniRV: AATGAAGGGGGGAGGAGTCAGA. A 295 bp fragment of the mitochondrial COI gene was amplified through PCR amplification using Bio-Rad T100 Thermal Cycler. The cycling conditions were 95 °C for 10 min, followed by 34 cycles of 95 °C for 45 s, 57 °C for 45 s, 72 °C for 45 s and a final step at 72 °C for 10 min.
COI amplicons were separated by 2.5% agarose gel electrophoresis followed by ethidium bromide staining and purified by elution following the Promega Wizard Purification Kit protocol. They were sequenced in outsourcing at Macrogen Korea using both COI forward and reverse primers. To compare results about 20% of samples were randomly repeated using CEQTM DTCS-Quick Start Kit (Beckman Coulter) and analyzed by means of capillary electrophoresis using an automated CEQ8000 DNA Analysis System (Beckman Coulter). Sequences were manually edited for sequential errors using MegaX software (Kumar et al. 2018) then they were aligned using ClustalW algorithm (Thompson et al. 1994). Species identity was determined based on a trimmed equal length by comparing each sequence in GenBank using the Basic Local Alignment Search Tool (BLAST) function. A similarity cutoff of ≥ 97% was used for species level identification for sequences submitted to the GenBank database.
2.4 Statistical analysis
After the alignment, new haplotypes were checked with all available sequences of Amphilius, Coptodon and Oreochromis in Genbank and analyzed with TCS software version 1.21 (Clement et al. 2000).
A Maximum Likelihood phylogenetic tree was obtained from the COI mtDNA sequences using Phylip software 3.0 (Guindon et al. 2010). Best substitution model was selected by Akaike Information Criterion (AIC) and branch support was performed by aLTR (Anisimova and Gascuel 2006). The phylogenetic tree was visualized by FigTree software (version 1.6).
3 Results
A final number of fifty-seven samples were successfully amplified for mtDNA barcoding out of 67 initially selected. In particular, 16 COI sequences of Amphilius from the museum collection and 41 of Tilapia (Table 1) were analyzed. Only 16 museum samples (3.0%) were successfully amplified with the COI barcoding from Ivanova et al. (2007) due to DNA degradation and fragmentation. Similarly, some of Tilapia samples were discharged due to DNA fragmentation as a consequence of very high temperatures during the field campaigns and no ice available.
Four different genera emerged from analysed sequences: Amphilius, Coptodon, Oreochromis and Tilapia (accession numbers from GenBank and taxonomic data are listed in Table 2). The final consensus length of all barcoded sequences was 684 bp. No stop codons, insertions o delections were observed. The frequency analysis of the nucleotide pairwise comparison showed that 426 of 684 (62.28%) sites were conserved in all samples, and the percentage of variable sites was 194 over 684 (28.36%). Informative parsimony sites were 171 (25%), and 23 (0.03%) singleton sites were present. The most representative tree model based on nucleotide substitutions was the HKY85 (Hasegawa-Kishino-Yano, 85). Twenty new haplotypes were described for the first time: 12 for genus Orechromis, 2 for Tilapia, and 2 for Coptodon among cichlids; 4 for Amphilius. Concerning single species the haplotype distribution was the following: A. uranoscopus (5), O. mossambicus (5), O. niloticus (6), O. karongae (1), C. rendalli (2), T. sparrmanii (2). The Nucleotides frequencies were: 0.26 (A), 0.31 (C), 0.16 (G), 0.27 (T).
Maximum Likelihood trees (Figs. 2, 3) was built with samples of this study together with COI sequences of Oreochromis, Coptodon (Fig. 2) and Amphilius genera (Fig. 3) from GenBank. As the best model of nucleotides substitution the GTR by AIC method was selected.
All the sequences from museum collection were subsequently amplified with COI mini-barcode and 30 samples out of 53 (56.6%) were successfully amplified for 198 bp (Table 3).
4 Discussion
The barcoding approach based on analysis of COI fragments is a useful methodology to study the taxonomy of freshwater fish and define new genetic lineages (Iyiola et al. 2018). The identification of native freshwater fish is necessary to delineate the evolutionary history of these species and detect new mitochondrial variants enriching the molecular diversity dataset of Southern Mozambique rivers. In this work a high number of newly detected sequences has been described for the first time revealing a widespread underestimation of the biodiversity of Cichlidae and Amphiliidae in this African country. Despite the limited number of analyzed sequences, due to sampling difficulties during field trips, a large number of new haplotypes witnesses the still existing gap of knowledge regarding freshwater fishes of Mozambique.
Although based on just one mitochondrial marker, the results highlight a large amount of molecular diversity in agreement with a recently published paper by Mazungula and Chakona (2021). These two authors discovered four new species of Amphilius in a nearby sampling site in Southern Mozambique. Despite the geographical closeness and similar habitats our experimental sequences of Amphilius clustered in a different node separated from the main cluster of A. uranoscopus described by Mazungula and Chakona (2021). However, whether or not this is a new species or an ecotype will have to be fully demonstrated in the future with further investigations. In any case, results highlight how understudied this genus is and its still undefined geographical range. It can be hypothesized that this taxonomic group in Mozambique more properly belongs to a species complex still unresolved. Nowadays, the coupling of traditional taxonomy based on morphological characteristic and molecular methods with the aim to detect divergent lineages witnessing different evolutionary histories is fundamental (Chakona et al. 2018; Thomson et al. 2015).
DNA barcoding is also an efficient tool for the identification of possible mismatch between species and incorrect morpho-identification. In fact, regarding the Cichlidae family, in this study different unresolved sequences and mismatches were found for Oreochromis sp., Tilapia and Coptodon genera (Fig. 2). More precisely, the genus Tilapia is undergoing revision and sequences of Tilapia guineensis obtained from GenBank cluster inside our group of Coptodon samples. A generalised difficulty in the tilapia taxonomy has already been demostrated by Sonet et al. (2019) that highlighted inconsistent species assigments between Coptodon and different similar genera inhabiting the Congo River due to undescribed species and unknown intraspecific variation. It is also noteworthy observing that increasing of big data to public databases has been associated with the increase of erroneous sequences that could be due to misidentified species and potential errors in sequencing (Shen et al. 2016). Particularly for the tilapia taxon, genetic studies can map existing species in the wild taking into account introduced fish and indicate the extent to which new species pose a danger in hybridization, genetic mutations and the extinction of native taxa (Cassemiro et al. 2018).
Results from mini-barcode DNA analysis has previously revealed the efficiency of this technique to prevent species fraud on commercial markets (Shokralla et al. 2015; Filonzi et al. 2021). Our similar approach also demonstrated the efficiency in the case of museum samples with damaged DNA. In fact, 56.6% of samples were successfully sequenced by mini-barcode versus 3% of reliable results obtained with the method of Ivanova et al. (2007).
In this work, the increasing occurrence of Oreochromis niloticus in natural habitats may be an indicator of how aquaculture is carried out in the country and may impact natural resources and endemic species. In this case, little or no information exist regarding the introduction of this North African species in Mozambique and genetic results open new perspectives in conservation biology of African fish considering the high reproductive potential and ecological plasticity of these invasive species connected to the fast generation of molecular variants.
References
Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55(4):539–552. https://doi.org/10.1080/10635150600755453
Bezeng BS, Van Der Bank HF (2019) DNA barcoding of southern African crustaceans reveals a mix of invasive species and potential cryptic diversity. PLoS One 14(9):1–12. https://doi.org/10.1371/journal.pone.0222047
Cassemiro ASF, Bailly D, Da Graça WJ, Agostinho AA (2018) The invasive potential of tilapias (Osteichthyes, Cichlidae) in the Americas. Hydrobiologia 817:133–154. https://doi.org/10.1007/s10750-017-3471-1
Chakona A, Kadye WT, Bere T, Mazungula DN, Vreven E (2018) Evidence of hidden diversity and taxonomic conflicts in five stream fishes from the Eastern Zimbabwe Highlands freshwater ecoregion. ZooKeys 768:69–95. https://doi.org/10.3897/zookeys.768.21944
Clement M, Posada D, Crandall KA (2000) TCS: A computer program to estimate gene genealogies. Mol Ecol 9:1657–1659. https://doi.org/10.1046/j.1365-294x.2000.01020.x
Decru E, Moelants T, De Gelas K, Vreven E, Verheyen E, Snoeks J (2016) Taxonomic challenges in freshwater fishes: a mismatch between morphology and DNA barcoding in fish of the north-eastern part of the Congo basin. Mol Ecol Resour 16(1):342–352. https://doi.org/10.1111/1755-0998.12445
Filonzi L, Chiesa S, Vaghi M, Nonnis Marzano F (2010) Molecular barcoding reveals mislabelling of commercial fish products in Italy. Intern Food Res Int 43(5):1383–1388. https://doi.org/10.1016/j.foodres.2010.04.016
Filonzi L, Vaghi M, Ardenghi A, Rontani PM, Voccia A, Nonnis Marzano F (2021) Efficiency of DNA mini-barcoding to assess mislabeling in commercial fish products in Italy: an overview of the last decade. Foods 10:1449. https://doi.org/10.3390/foods10071449
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010)New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59(3):307–321. https://doi.org/10.1093/sysbio/syq010
Guindon S, Gascuel O (2003) PhyML: a simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52(5):696–704. https://doi.org/10.1080/10635150390235520
Hajibabaei M, Smith MA, Janzen DH, Rodriguez JJ, Whitfield JB, Hebert PDN (2006) A minimalist barcode can identify a specimen whose DNA is degraded. Mol Ecol Notes 6:959–964. https://doi.org/10.1111/j.1471-8286.2006.01470.x
Ivanova NV, Zemlak TS, Hanner RH, Hebert PD (2007) Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes 7(4):544–548. https://doi.org/10.1111/j.1471-8286.2007.01748.x
Iyiola OA, Nneji LM, Mustapha MK, Nzeh CG, Oladipo SO, Nneji IC, Okeyoyin AO, Nwani CD, Ugwumba OA, Ugwumba AAA, Faturoti EO, Yyu W, Chen J, Wang WZ, Adeola AC (2018) DNA barcoding of economically important freshwater fish species from north-central Nigeria uncovers cryptic diversity. Food Res Int 8(14):6932–6951. https://doi.org/10.1002/ece3.4210
Kumar S, Stecher G, Li M, Knyaz C, Tamura K, Battistuzzi FU (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Mazungula DN, Chakona A (2021) An integrative taxonomic review of the Natal mountain catfish, Amphilius natalensis Boulenger 1917 (Siluriformes, Amphiliidae) with description of four new species. J Fish Biol. https://doi.org/10.1111/jfb.14714
Mojekwu TO, Cunningham MJ, Bills RI, Pretorius PC, Hoareau TB (2021) Utility of DNA barcoding in native Oreochromis species. J Fish Biol 98(2):498–506. https://doi.org/10.1111/jfb.14594
Pereira LHG, Hanner R, Foresti F, Oliveira C (2013) Can DNA barcoding accurately discriminate megadiverse Neotropical freshwater fish fauna? BMC Genet 14:1–14. https://doi.org/10.1186/1471-2156-14-20
Russell DJ, Thuesen PA, Thomson FE (2012) A review of the biology, ecology, distribution and control of Mozambique tilapia, Oreochromis mossambicus (Peters 1852) (Pisces: Cichlidae) with particular emphasis on invasive Australian populations. Rev Fish Biol Fish 22(3):533–554. https://doi.org/10.1007/s11160-011-9249-z
Schmidt RC, Bart HL, Jr N, Nyingi WD (2015) Two new species of African suckermouth catfishes, genus Chiloglanis (Siluriformes: Mochokidae), from Kenya with remarks on other taxa from the area. Zootaxa 4044(1):45–64
Shen Y, Guan L, Wang D, Gan X (2016) DNA barcoding and evaluation of genetic diversity in Cyprinidae fish in the midstream of the Yangtze River. Ecol Evol 6(9):2702–2713. https://doi.org/10.1002/ece3.2060
Shokralla S, Hellberg R, Handy S, King I, Hajibabaei M (2015) A DNA mini-barcoding system for authentication of processed fish products. Sci Rep 5:15894. https://doi.org/10.1038/srep15894
Sonet G, Snoeks J, Nagy ZT, Vreven E, Boden G, Breman FC, Decru E, Hanssens M, Ibala ZA, Jordaens K, Mamonekene V, Musschoot T, Van Houdt J, Van Steenberge M, Lunkayilakio WS, Verheyen E (2019) DNA barcoding fishes from the Congo and the lower guinean provinces: assembling a reference library for poorly inventoried fauna. Mol Ecol Resour 19(3):728–743. https://doi.org/10.1111/1755-0998.12983
Staats M, Arulandhu AJ, Gravendeel B, Holst-Jensen A, Scholtens I, Peelen T, Prins TW, Kok E (2016) Advances in DNA metabarcoding for food and wildlife forensic species identification. Anal Bioanal Chem 408(17):4615–4630. https://doi.org/10.1007/s00216-016-9595-8
Sultana S, Ali ME, Hossain MM, Naquiah N, Zaidul I (2018) Universal mini COI barcode for the identification of fish species in processed products. Food Res Int 105:19–28. https://doi.org/10.1016/j.foodres.2017.10.065
Thompson JD, Higgs DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties, and weight matrix choice. Nucleic Acids Res 22:4673–4680
Thomson AW, Page LM, Hilber SA (2015) Revision of the Amphilius jacksonii complex (Siluriformes: Amphiliidae), with the descriptions of five new species. Zootaxa 3986:61–87
Waugh J (2007) DNA barcoding in animal species: progress, potential and pitfalls. BioEssays 29(2):188–197. https://doi.org/10.1002/bies.20529
Whitfield JB (1999) Destructive sampling and information manage-ment in molecular systematic research: an entomological perspec-tive. In: Byers S, Metsger D (eds) Managing the modern herbarium: an interdisciplinary approach. Society for Preserva-tion of Natural History Collections and Royal Ontario Museum, p 384
Yáñez JM, Joshi R, Yoshida GM (2020) Genomics to accelerate genetic improvement in tilapia. Anim Genet 51(5):658–674. https://doi.org/10.1111/age.12989
Acknowledgements
This research was financially supported by the Italian Agency for Development Cooperation and is part of the project “Formazione in biodiversità e biotecnologia per la conservazione ambientale e lo sviluppo sostenibile—AID 11096”. The work has benefited from the equipment and framework of the COMP-HUB Initiative, funded by the ‘Departments of Excellence’ program of the Italian Ministry for Education, University and Research (MIUR, 2018–2022). The authors are particularly grateful to Prof. Mauro Maria Colombo for his management skills and coordination of the entire project, and the director of the Natural History Museum of Maputo Mrs. Lucilia Chuquela for administrative support.
Funding
Open access funding provided by Università degli Studi di Parma within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This paper belongs to a Topical Collection originated from a long scientific collaboration between Mozambican and Italian universities promoted by the Italian Agency for Development Cooperation.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ferrari, C., Tovela, E., Taviani, E. et al. DNA barcoding to assess species identification in museum samples of Amphiliidae and natural samples of Cichlidae from Southern Mozambique. Rend. Fis. Acc. Lincei 33, 713–720 (2022). https://doi.org/10.1007/s12210-022-01098-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12210-022-01098-1