Abstract
In this research, eight kiwi fruit genotypes (six hardy kiwis (Actinidia arguta and their hybrids), one of Actinidia deliciosa ‘Hayward,’ and one of Actinidia eriantha ‘Bidan’ were examined and compared by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), Fourier transform infrared (FT-IR) spectra, and nuclear magnetic resonance (NMR) spectroscopy. Proteins were extracted from lyophilized fruits, flesh with seeds, grinded seeds, and singular seeds and then separated by SDS-PAGE. Matrix similarity and dendrogram was generated using Nei coefficient and Unweighted Pair Group Method with Arithmetic mean (UPGMA) algorithm. Based on protein patterns, Actinidia species were clearly distinguishable, whereas differences between hardy kiwi fruit cultivars were minor or nondetectable. The electrophoretical separations were able to distinguish a half of hardy kiwi fruit cultivars, so cluster analysis revealed a limited number of cultivar groups. Intervarietal polymorphism was low and this affected the results of similarity analysis. One distinct cluster, composed of two pairs of cultivars and identical by protein patterns, was obtained. Cultivars ‘Ananasnaya’ and ‘Weiki,’ according to the morphological description, were similar. Oppositely, ‘M1’ cultivar significantly differed from other hardy kiwi cultivars by densitometrical bands intensity. All examined singular seeds of ‘Ananasnaya’ cultivar possessed identical protein patterns. The protein patterns of ‘Bingo’ and ‘Ananasnaya’ hardy kiwi fruits harvested in 2011 and 2013 were identical. Three weeks storage after harvest did not affect the protein composition of these cultivars. FT-IR and NMR spectrum of hardy kiwi fruits were presented and compared with ‘Hayward’ and ‘Bidan’ and showed slight differences in comparison with the protein profiles. SDS-PAGE is more applicable than FT-IR and NMR for comparison of different kiwi fruit cultivars. The used analytical methods can be applied to any food analysis in order to distinguish the main compounds and to present fingerprints of different cultivars.

A, Actinidia arguta kiwi fruit cultivars; B, Actinidia deliciosa kiwi fruit ‘Hayward’; C,Actinidia eriantha kiwi fruit ‘Bidan’; D, Electrophoretical patterns of proteins extracted with urea bufferfrom: 1, ‘Bingo’; 2, ‘M1’; 3, ‘Ananasnaya’; 4, ‘Weiki’; 5, ‘Jumbo’; 6, ‘Geneva’; 7, ‘Hayward’; 8,‘Bidan’; E, FT-IR spectra of extracted polyphenols. Curves from the top kiwi fruit ‘Ananasnaya,’‘Bingo,’ ‘M1’; F, 1H-NMR spectra from the top of DMSO extracts of ‘Bidan’; ‘Hayward’;‘Ananasnaya’.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Actinidia Lindl belongs to Actinidiaceae family and comprises 66 species and 118 taxa. Acinidies are dioecious or, rarely, monoecious perennial outcrossing vines. All members of the genus can be characterized by the variation in ploidy level including diploids (2n = 58), tetraploids (2n = 116), and hexaploids (2n = 174). The taxonomy of Actinidia, based on the morphological characters, is difficult due to the significant morphological variations and existing reticulated intraspecific and interspecific structures within the genus. These structures have occurred probably by natural crossing (Huang et al. 2000; Huang et al. 2002). In the past 10 years, the number of kiwi fruit cultivars commercially selected has significantly increased. Essentially, all these cultivars were derived just from 4 of 66 species, which are present within the Actinidia genus. The most popular from Actinidia species on the fruit market are Actinidia deliciosa (A. Chev.) C. F. Liang et A.R. Ferguson and closely related Actinidia chinensis Planch. A. deliciosa and A. chinensis fruits are typically large, 60–100 g, and green- or red-flashed. The predominant cultivar of A. deliciosa is ‘Hayward.’ The health benefits of kiwi fruits are attributed in part to bioactive compounds and possessed high antioxidant capacity, although their value can be changed during ripening (Park et al. 2012a, 2015a). Different extraction procedures and antioxidant assays were used for characterization of their bioactive compounds (Park et al. 2012a, b, 2013, 2015a, b, c). Nowadays, kiwi fruits are cultivated mainly in East Asia, on 100,000 ha, with annual production of about one million tons. Lately, in Europe, the cultivation of other Actinidia species such as Actinidia arguta (Sieb. Et Zuch Planch. Ex Miq) and Actinidia eriantha is becoming popular. A. arguta are also called hardy kiwi fruits, mini kiwis. There are many varieties of hardy kiwi fruit cultivated worldwide, and some of them are cultivated in Poland, including hybrids (Latocha and Olszewska-Kaczyńska 2003; Latocha and Krupa 2007). Hardy kiwis are smaller than traditional kiwi fruits (about 6–14 g), hairless, sweet and they vary in color and shape. In contrary to traditional kiwi fruits, they have smooth edible skin. Hardy kiwi fruits are frost resistant up to −30 °C and can be cultivated in countries with cold climate. Additionally, hardy kiwis are nearly free of pests and diseases, and are perfectly suitable for ecological cultivation. Hardy kiwi fruits, harvested unriped (about 6.5–7 °Brix) can be stored in refrigerator up to 8 weeks, while widely consumed kiwi fruits can be kept up to half-year (Latocha and Krupa 2007). A. eriantha fruits are small, hairy, and potentially easy to peel. ‘Bidan,’ a new cultivar of A. eriantha, was selected in South Korea. The average fruit weight is 25 g. The cultivar ‘Bidan’ has high content of vitamin C and antioxidative activity compared to the A. deliciosa cultivar ‘Hayward’ (Jo et al. 2007; Park et al. 2013; Li et al. 2014; Park et al. 2015c). Three major allergenic proteins of kiwi fruits, such as kiwellin (Tamburrini et al. 2005; Tupo et al. 2008; Maddumage at al. 2013), thaumatin-like protein (TLP) (Maddumage at al. 2013), pectin methylesterase inhibitor (Ciardello et al. 2004), and actinidin (Ciardello et al. 2004; Nishyama 2007; Maddumage et al. 2013) have been isolated, identified, and characterized with the use of Western blotting. The increase of a number of cultivated both traditional kiwi fruits and hardy kiwi varieties and in a consequence of increase of nursery material amount, fruits and seeds trade, implicates a necessity of developing reliable methods of genotypes identification in collections. Identification of genotypes on the basis of morphological and biochemical characters, verification of their identity, and in the case of seed trade evaluation of seed genetic purity require rapid analytical methods. Up to now, kiwi fruit cultivars were distinguished by leaf isozyme polymorphism on the starch gels. The level of intervarietal polymorphism on the basis of protein bands pattern was low, and only a part of used isoenzyme systems was suitable for this purpose. About 500 plants of 20 kiwi taxa, followed by starch gel electrophoresis (SGE) and separation of isozymes, were successfully examined, using 16 enzyme systems. Based on SGE data, Unweighted Pair Group Method with Arithmetic mean (UPGMA) dendrogram was generated and grouped in different clusters: three A. arguta forms, three A. deliciosa varieties, and one of A. eriantha genotype (Testolin and Ferguson 1997). Some improvement was achieved by the use of biochemical markers isolated from leaves, on the basis of sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) protein patterns. Eleven A. deliciosa varieties were partially distinguished on acrylamide gels (Zhu and Lawes 1990). The most significant improvements of understanding the phylogenetic relationships (on the example of 40 taxa, including 31 species) were molecular (RAPD) markers obtained from leaf material. Twenty-two oligonucleotide primers were used and 204 DNA bands were obtained, of which 192 (92 %) were polymorphic. The average similarity between different species was 0.28, and between varieties of the same species was 0.54, respectively. With the use of dendrogram, the taxonomy of the genus could be verified by grouping taxa (species, varieties) into clusters. Actinidia taxa were grouped into eight clusters. Kiwi fruits A. deliciosa, A. eriantha (classified into Stellatae Li section), and hardy kiwis (belonged to Leiocarpae) were grouped into different clusters—fourth, second, and seventh, respectively (Huang et al. 2002). The difficulties in distinguishing kiwi fruit varieties on the basis of leaf isoenzyme characters and leaf biochemical markers were an inspiration for us to develop new analytical biochemical markers, both of fruits and seeds, suitable for distinguishing A. arguta varieties and, additionally, for discriminating ‘Hayward’ and ‘Bidan’ cultivars. There are known data about quantitative differences found in some kiwi cultivars, based on their chemical compounds such as actinidin and kiwellin activity (Ciardello et al. 2004; Nishyama 2007; Maddumage et al. 2013). Knowledge about the identity characteristics as biochemical markers that can be used for discrimination of kiwi fruit genotypes is limited (Zhu and Lawes 1990; Testolin and Ferguson 1997). We were interested to find out whether the quality of protein patterns derived from fruits will be sufficient for grouping kiwi fruit genotypes in clusters, with the use of significantly lower work loading than those used for starch gel electrophoresis (SGE). There is an increased requirement by plant breeders and food chemists to develop reliable methods of identity evaluation of new kiwi fruit genotypes (Latocha and Krupa 2007). Any publications that show differences between kiwi species based on the biochemical markers, as an electrophoretical pattern of proteins, suitable for discriminating A. arguta varieties too, are not found. To the best of our knowledge, no results of applying protein markers derived from seeds for distinguishing kiwi fruit cultivars and potentially suitable for evaluation of genetic variation and purity of seed samples do exist. We have supposed that it would be possible to do this by analysis of electrophoretic patterns of proteins extracted from singular seeds. As far as we know, there are no published reports on the subject of possible changes in the protein profile collected from horticultural plants such as kiwi fruits, harvested in different years. We have tried to answer these questions by comparing the electrophoretic patterns of proteins of two hardy kiwi fruit cultivars ‘Bingo’ and ‘Ananasnaya’ harvested in 2011 and 2013, and then stored for 3 weeks. We also evaluated the possibility of using the attenuated total reflectance accessory (ATR-FT-IR) and nuclear magnetic resonance (NMR) spectroscopy for comparison of all studied samples (He et al. 2007; Anastasiadi et al. 2009; Lopez-Sánchez et al. 2010; Capitani et al. 2013a).
Materials and Methods
Chemicals
SDS, β-mercaptoethanol, acrylamide, bis-acrylamide, tris (hydroxymethyl) aminomethane (Tris), glycine, urea, dithiothreitol (DTT), and Coomassie Brillant Blue R-250 were purchased from Sigma Chemical Co., St. Louis, MO, USA. All reagents were of analytical grade. Deionized and distilled water was used throughout.
Serum albumin Bovine 67 kDa, ovalbumin Chicken egg 45 kDa, carbonic anhydrase Bovine erythrocytes 29 kDa, trypsin inhibitor Soybean 21 kDa, alpha-lactalbumin 14.4, lysozyme Chicken egg 12.5 kDa and aprotinin Bovine lungs 6.5 kDa were purchased from Serva Electrophoresis GmbH, Heidelberg, Germany, and used as molecular markers.
Kiwi Fruit Samples
Hardy kiwis (A. arguta) plants, grown on sandy loam soil, were collected in 2011 and 2013, from the experimental ecological field of the Department of Environmental Protection, Warsaw University of Life Sciences, Poland. Fruits of the following cultivars: ‘Bingo’ (hybrid of A. arguta and Actinidia purpurea), ‘M1’ (select arguta), ‘Ananasnaya,’ ‘Weiki,’ ‘Jumbo,’ and ‘Geneva’ were picked up at their eating maturity of the fruit softness. Additionally, fruits of ‘Bingo’ and ‘Ananasnaya’ were collected at the harvesting maturity (7 °Brix) and stored for 3 weeks after harvest (1 °C; 95 % RH) and were ripened in presence of ethylene (500 ppm) to eating maturity stage. Fruits were first washed under running water and freezed and then were freeze-dried together with the peel in Company ELENA, Kokanin 86, Poland. Kiwi fruit cultivars ‘Hayward’ (A. deliciosa) and ‘Bidan’ (A. eriantha) were grown in organic system without using any pesticides or herbicides, in an orchard Heanam County, Jeonnam Province, South Korea, in 2011 and 2013. Samples were cleaned with water and the peeled fruits were weighed, chopped, and then lyophilized.
Preparation of Protein Samples for Electrophoresis
Proteins of A. arguta cultivars were extracted from the following: (i) fruit lyophilizates, (ii) flesh with seeds, and (iii) seeds. Seeds were isolated from 20 (‘M1’) or 10 (other cultivars). In the case of A. deliciosa and A. eriantha cultivars, proteins were extracted only from lyophylizates.
Protocols of Proteins Extraction
From Fruit Juice
The fruits were thawed, then 1.5 mL of filtrate was centrifuged at 15,000×g for 20 min. The sediment was dissolved in a volume of 41.5 μL buffer (0.5 M Tris-HCl, pH 8.3) and mixed with 41.5 μL of Sample Buffer (1X, Sigma).
From Fruit Lyophilizate
The proteins were extracted from lyophilizate powder with urea buffer with the following formula: 2.6 M urea solution, 5 % DTT, 0.22 M SDS, and 0.02 % bromophenol blue in ratio 60 mg of powder/500 μL urea buffer.
From Bulk Samples
Seeds (100 and 200 mg) were crushed into powder and then were mixed with the urea buffer in the ratio 100 and 200 mg/1000 μL urea buffer.
From Singular Seeds
Seeds were crushed and then mixed with 20 μL of urea buffer. Before application on polyacrylamide gel, all protein samples were boiled for 3 min and then centrifuged at 18,000×g for 7 min.
Protein Electrophoresis
The protein supernatants were loaded on the polyacrylamide gels in a volume of 10 μL. The SDS-PAGE was carried out according to the common formula of Laemmli (1970), using a Hoeffer SE-600 vertical unit (Hoeffer Scientific Instruments, San Francisco, CA, USA), and the resolving gel was 13.7 % T and 1.7 % C, and the stacking gel was 3.8 % T and 1.8 % C. The gel size was 140 × 160 × 1.5 mm. After run gels were fixed for 30 min in 15 % solution of TCA, then they were rinsed in water and stained in Coomassie Blue R-250 solution at room temperature overnight and then destained.
Analysis of Similarity (Homology) Between Cultivars
The banding patterns of cultivar profiles from the electrophoregram were scored as bands for presence or absence of the band to estimate occurrence or absence of intervarietal polymorphism. The computer software BIO GENE (Vilber Lourmat, France, ver. 99.03) was used to calculate the Nei and Li (1979) genetic similarity coefficients (GS) and for generating the similarity matrix for all examined pairs of cultivars. Similarity coefficients (also called Dice) = 2nxy (nx + ny), where nx and ny are the number of bands in the lane x and in the lane y, respectively, and nxy is the number of share bands between the two lanes. The dendrogram was generated from the similarity (homology) values and using UPGMA algorithm.
The Densitometry Analysis of Gel Protein Bands
This analysis was performed with the use of BioGene software v. 11.9 Vilbert Lourmat, France. The densitometry profiles of proteins of all examined cultivars were obtained as curves of peaks height of maximum intensity on a range of 256 gray levels.
Fourier Transform Infrared Spectra of Polyphenols
Total phenols in the investigated fruit extracts were studied by IR spectroscopy. A Nicolet iS 10 Fourier transform infrared (FT-IR) Spectrometer (ThermoScientific Instruments LLC, Madison, WI, USA), with the smart iTRTM attenuated total reflectance (ATR) accessory was used to record IR spectra (Park et al. 2015a, c; Lopez-Sanches et al. 2010).
1H NMR Spectroscopy
Each extract of kiwi fruit sample was dissolved in 700 μL of dimethyl sulfoxide (DMSO). Extraction was done also with methanol/chloroform/water at 2:2:1 volumetric ratio. Sample was kept at 4 °C for 1 h and then centrifuged for 20 min at 11,000×g at 4 °C. The upper hydroalcoholic phase was carefully separated and dried under a N2 flow. The dried phase was stored at −80 °C until the NMR analysis (Capitani et al. 2013b). All NMR experiments were recorded on Bruker 500 NMR spectrometer equipped with a 5-mm PABBO BB-probe head (499.953 for 1H shifts and 125.713 for 13C shifts) at 25 °C. NMR data processing was performed using MestReNova software (Anastasiadi et al. 2009; Capitani et al. 2013a).
Results and Discussion
The Electrophoretical Spectra of Kiwi Fruits Proteins
The investigated varieties of kiwi fruits are presented on Fig. 1a–c. The SDS-PAGE spectrum from fruit juices of six hardy kiwi varieties (Fig. 2a) consists of six to seven bands and only one of them (‘M1’) was distinguished from other hardy kiwi varieties (Fig. 2a, arrow a). Extracts from fruit lyophilizates revealed significant differences in the protein patterns of three kiwi fruit species. The electrophoretic spectrum of hardy kiwis consisted of 13–14 bands of different intensities with molecular weights of 14–52 kDa, while A. deliciosa and A. eriantha were composed only of some bands (Fig. 2b, arrows a–c and h showed differences between kiwi fruit species). The major protein bands of hardy kiwi cultivars were in the ranges from 17 to 22 kDa and 28–36 kDa. In ‘Hayward,’ the major bands were at 22, 24, and 32 kDa. It was possible to obtain only six sharp bands in the protein pattern of ‘Bidan,’ using urea buffer. One of the bands, characterized by molecular weight of 23 kDa, was more intensive by the amount of protein stained on the gel and comparable to bands intensity in patterns of other cultivars. Hardy kiwi fruits varieties were nearly monomorphic by protein patterns, and the intervarietal differences were small or any differences were not detected (Fig. 2b, arrows d–g, j, and k showed differences among four of six varieties examined). ‘M1’ cultivar differs from others by the presence of bands that are shown as d, e, j, k, but in ‘Ananasnaya,’ ‘Weiki,’ and ‘Jumbo’ the number of differentiating bands is limited to one (shown as f and g bands). Four groups of hardy kiwi fruit cultivars can be distinguished on the basis of protein spectrum: the first and the second were two pairs of identical cultivars: ‘Bingo’ and ‘Geneva,’ and ‘Ananasnaya’ and ‘Weiki’. ‘M1’ and ‘Jumbo’ clearly differ from other cultivars. Especially, the protein spectrum of ‘M1’ was distinct from other patterns due to the bands: 17 kDa (Fig. 2b, arrow d) and 52 kDa (Fig. 2b, arrow k), while no band of 31 kDa common for other hardy kiwi varieties was detected (Fig. 2b, arrow e). In the case of A. deliciosa and A. eriantha cultivars (‘Hayward’ and ‘Bidan’), three distinct interspecies differences were found (Fig. 2b, arrows b, c and h).
a Electrophoretical patterns of proteins extracted with Tris-HCl and sample buffers from fruit juices of six hardy kiwi cultivars and separated by SDS-PAGE. Arrow a indicates band that distinguish ‘M1’ from other cultivars. 1, ‘Bingo’; 2, ‘M1’; 3, ‘Ananasnaya’; 4, ‘Weiki’; 5, ‘Jumbo’; 6, ‘Geneva’. b Electrophoretical patterns of proteins extracted with urea buffer from fruit lyophilizate of hardy kiwis and kiwi fruit varieties and separated by SDS-PAGE. 1, ‘Bingo’; 2, ‘M1’; 3, ‘Ananasnaya’; 4, ‘Weiki’; 5, ‘Jumbo’; 6, ‘Geneva’; 7, ‘Hayward’; 8, ‘Bidan’. Arrows a–c indicate band patterns differences between kiwi species, arrows d–g, j, and k indicate bands that distinguish particular hardy kiwi cultivars from each other. Arrows b, c, and h indicate major differences of band patterns between ‘Hayward’ and ‘Bidan.’ c Electrophoretical patterns of proteins extracted with urea buffer from fruit lyophilizates of hardy kiwi fruit cultivars ‘Bingo’ and ‘Ananasnaya,’ collected in 2011 and 2013 and stored for 3 weeks. 1, ‘Ananasnaya,’ 2011 harvest; 2, ‘Ananasnaya,’ 2013 harvest; 3, ‘Ananasnaya,’ stored; 4, ‘Bingo,’ 2011 harvest; 5, ‘Bingo,’ 2013 harvest; 6, ‘Bingo,’ stored. Molecular weight markers for a–c: serum albumin Bovine 67 kDa, ovalbumin Chicken egg 45 kDa, carbonic anhydrase Bovine erythrocytes 29 kDa, trypsin inhibitor Soybean 21 kDa, lysozyme Chicken egg 12 kDa and aprotinin Bovine lungs 6 kDa
Protein Patterns of ‘Bingo’ and ‘Ananasnaya’ Cultivars
In protein patterns of two cultivars, collected in 2011 and 2013, no changes were determined (Fig. 2c). Storage had no effect on fruit protein patterns.
Electrophoresis of Hardy Kiwi Proteins Extracted from 100 to 200 mg Seed Samples
Electrophoresis of each of examined cultivars revealed the same patterns (Fig. 3a). The protein spectrum of bulk samples consisted of 24–26 bands with three zones of very intensive stained bands: 16–22, 29–33, and about 50 kDa. Two varieties ‘Jumbo’ and ‘Anananasnaya’ slightly differ from other cultivars (Fig. 3a, arrows a and b). Electrophoretic spectra of proteins extracted from singular seeds of six hardy kiwi cultivars consisted of 18–20 bands, but revealed slightly higher polymorphism of patterns than those obtained from “bulk samples.” Slight intervarietal differences were detected in patterns of ‘M1,’ ‘Jumbo,’ and ‘Bingo’ cultivars (Fig. 3a, arrows c–e, respectively).
a Electrophoretical patterns of proteins extracted with urea buffer both from 100 and 200 mg weighted seed samples and from singular seeds of hardy kiwi cultivars and then separated by SDS-PAGE. Arrows a and b indicate bands that distinguish ‘Jumbo’ and ‘Ananasnaya’ cultivars, based on protein patterns derived from bulk sample materials. Arrows c–e indicate bands that distinguish cultivars ‘M1,’ ‘Jumbo,’ and ‘Bingo’ from each other, based on protein patterns of singular seeds. 1–12—protein patterns of bulk samples, 13–18—protein patterns of singular seeds. 1, ‘Bingo,’ 100 mg; 2, ‘Bingo,’ 200 mg; 3, ‘M1,’ 100 mg; 4, ‘M1,’ 200 mg; 5, ‘Ananasnaya,’ 100 mg; 6, ‘Ananasnaya,’ 200 mg; 7, ‘Weiki,’ 100 mg; 8, ‘Weiki,’ 200 mg; 9, ‘Jumbo,’ 100 mg; 10, ‘ Jumbo,’ 200 mg; 11, ‘Geneva,’ 100 mg; 12, ‘Geneva,’ 200 mg; 13, ‘Bingo,’ sing.; 14, ‘M1,’ sing.; 15, ‘Ananasnaya,’ sing.; 16, ‘Weiki,’ sing.; 17, ‘Jumbo,’ sing.; 18, ‘ Jumbo,’ sing. Abbreviations: bulk sample 100 mg (100 mg), bulk sample 200 mg (200 mg), singular seeds (sing.). b Electrophoretical patterns of proteins extracted with urea buffer from singular seeds of hardy kiwi cultivar ‘Ananasnaya’ and separated by SDS-PAGE. 1 to 22—singular seeds of ‘Ananasnaya.’ Molecular weight markers for a, b: serum albumin Bovine 67 kDa, ovalbumin Chicken egg 45 kDa, carbonic anhydrase Bovine erythrocytes 29 kDa, trypsin inhibitor Soybean 21 kDa and alpha-lactalbumin 14 kDa
Electrophoretic Spectra of Proteins Extracted From Singular Seeds of Hardy Kiwis
‘Ananasnaya’ cultivar was identical (Fig. 3b).
Statistical Analysis
A low level of polymorphism between protein patterns of hardy kiwi cultivars has been confirmed. A low level of polymorphism determined high level of genetic similarity between patterns-lanes of particular cultivars. Value of 100 % similarity was found for two pairs of cultivars: ‘Bingo’ and ‘Geneva,’ and ‘Anananasnya’ and ‘Weiki’ (similarity 100 % was shown on matrix as a coefficient 1.00, Fig. 4a). These cultivars pairs represented identical lanes. It means that from this point of identity, such genotypes were the same. Coefficient similarities of other pairs of hardy kiwi fruit cultivars were lower, but not lower than 0.67 (Fig. 4a). Coefficient similarities between ‘Hayward’ and ‘Bidan’ cultivars were lower and estimated as 0.57. These kiwi fruit genotypes differed significantly from hardy cultivars, showing the level of similarity varying from 0.29 to 0.67 (Fig. 4a). Dendrogram analysis of cultivars was not able to group them distinct clusters, except a cluster of two pairs of identical cultivars and, partially, cluster was generated of ‘Hayward’ and ‘Bidan’ (Fig. 4b). The densitometry analysis of protein profiles has generated curves-lanes partially “covered” each other (Fig. 4c). It means that in the course of densitometry lanes that were composed of peaks, quanititative differences by height of peaks were weak or no differences were shown, especially those that represented A. arguta cultivars. Any significance in a feature of peaks height which caused by lack of the differences in the presence of bands or their intensities was not estimated. It was impossible to distinguish A. arguta lanes on the basis of the densitometry lanes of particular cultivars, except ‘M1’. This corresponded with an evidence of low protein polymorphism of A. arguta cultivars. Such phenomenon was in other parts of curves in major peaks. Maximum height was reached for lane 2 (that represented ‘M1’ cultivar of A. arguta)—12,278,143. Heights of other cultivars were slightly lower. The densitometry curve of ‘Bidan’ cultivar was nearly “flat” and differed from other curves. As it was shown above, two protocols were used for extraction of total proteins (or urea-soluble proteins), of whole fruits to find a rapid, reproducible method of protein electrophoresis that could be used initially for A. arguta cultivars and then for other kiwi fruit species. Proteins were extracted in the presence of high level of DTT to ensure complete reduction of proteins. The use of buffer sample solution for extraction of A. arguta kiwi juice proteins was not promising, until the new extraction buffer for fruit lyophilizate was applied and a small progress was achieved. We have achieved a progress in ability of protein extraction from kiwi tissues (fruit lyophylizate, seeds) by the use of urea buffer. Our results are in line with Vergara-Barberan et al. (2015), who improved the protein extraction from olive leaves using an enzyme-assisted protocol with a cellulase enzyme. Different parameters that affect the extraction process, such as the influence and amount of organic solvent, enzyme amount, pH, and extraction temperature and time, were optimised. The improvement of electrophoregrams for the practice purposes was an increase of the number of bands detected on the electrophoregrams (up to 22–24 bands for seeds proteins). Other researchers have obtained a seed protein pattern for kiwi fruit which consisted only of 11 bands (Miraghaee at al. 2011; Miraghaee at al. 2012). Common opinion among seed scientists is that seed proteins are especially suitable for the aim of characterization and distinguishing of crop cultivars (Cooke 1989). We were interested whether this opinion can be considered as common one for seeds of fruit plants too. Unfortunately in the case of hardy kiwis, the level of the seed protein intervarietal polymorphism has remained relatively low, and also of limited suitability for this purpose. According to molecular taxonomy of genus Actinidia, molecular similarity between kiwi fruit species was approximately 0.28. Three examined in this study kiwi fruit species were classified to different clusters of the dendrogram (Huang at al. 2002). Our results have confirmed significant differences in the biochemical identity between three kiwi fruit species. We have confirmed data provided by others (Tamburrini et al. 2005; Miraghaee et al. 2011) that the most distinct bands were in the range from 20 to about 30 kDa, and particularly protein band of 30 kDa was identified by N-terminal amino acid sequencing as actinidin. Western blotting analysis allowed to find actinidin band at ~24 kDa in protein spectrum of A. deliciosa (among them ‘Hayward’ cv.), A. chinensis, and A. arguta cultivars. Band of 24 kDa was not detected in A. eriantha cultivars. This supported our data that protein pattern of A. eriantha cultivars was composed of lower number of bands than were detected in protein patterns of ‘Hayward’ and A. arguta cultivars (Maddumage et al. 2013). As it was shown previously, actinidin was the most abundant protein in fruit and leaves of kiwi fruit (Afshar-Mohammadian et al. 2011; Testolin and Ferguson 1997; Miraghaee et al. 2011), but three of A. arguta cultivars contained higher levels of actinidin than ‘Hayward’ (Afshar-Mohammadian et al. 2011; Nishyama 2007). Only trace amount of actinidin (25 kDa band) was found in juice of some kiwi fruit varieties, and intensity of actinidin band was the same as other bands of protein patterns. In the case of other varieties, intensity of actinidin band was higher (Nishyama 2007). For kiwellin, a band at ~20 kDa was observed in A. deliciosa, A. arguta, A. chinensis, and A. eriantha cultivars. TLP proteins (band at ~22 kDa) in A. eriantha occurred only in traces or completely in no detectable amount of protein (Maddumage et al. 2013). Our results showed that the coefficient intervarietal (A. arguta) similarity was higher than 0.67. This confirmed the conclusions of other researches that genetic similarity between botanical varieties of particular species of Actinidia was relatively high (0.54 in average) and probably was lower than those of cultivated ones. Molecular markers analysis also supported our data that kiwi fruit cultivars, including botanical varieties, were genetically similar. For example, RAPD markers allowed grouping of botanical varieties into a singular cluster. The forms of A. deliciosa var. chlorocarpa, A. deliciosa var. deliciosa, and A. deliciosa with A. chinensis var. chinensis were classified into two clusters, but nearby on the dendrogram (Huang et al. 2002). Similarly, clustering based on SGE method allowed to group A. arguta cultivars in one cluster, with the level of similarity between cultivars higher than 0.7 (Testolin and Ferguson 1997). And indeed, a factor of high genetic similarity between cultivars examined by us has not allowed to distinguish all A. arguta varieties on the basis of proteins patterns obtained from fruit and seeds tissues. Although, all examined varieties of A. arguta origin from different countries [‘Jumbo,’ ‘Weiki,’ and ‘Geneva’ were selected in Italy, Germany, and USA, respectively; ‘M1’ and ‘Bingo’ are hybrid varieties selected in Poland], the protein polymorphism detected on gels was low. We have obtained interesting data that ‘Weiki’ and ‘Ananasnaya’ (unknown origin) plants, according to the morphological description, were similar varieties (Latocha and Krupa 2007). Protein patterns isolated from juice and seeds became undistinguished. It should be emphasized that distinguishing of A. deliciosa varieties on the basis of isoenzyme features was difficult (Testolin and Ferguson 1997). Only a small improvement in cultivars discrimination was achieved by high-resolution polyacrylamide gradient gels. One of four examined hardy kiwi cultivars was clearly distinct on gel from others (Maddumage et al. 2013). This supported our suggestions of low genetic variability among hardy kiwi genotypes. ‘Hayward’ and ‘Bidan’ cultivars can be easily distinguished by protein patterns, although the number of bands detected on gels was low, especially in ‘Bidan.’ The explanation of these results can be that both genotypes are classified to different genetic pools of materials. Nevertheless, an increase of detected distinct bands on gels was obtained, but the number of bands that characterize A. eriantha cultivars remained lower than other examined kiwi fruits (Maddumage et al. 2013). Densitometry analysis showed a flat curve of densitometry peaks, which was typical for ‘Bidan.’ According to this feature, ‘Bidan’ differed from other examined cultivars. On the other hand, peaks of ‘M1’ cv. were higher than others and it can be connected with the highest antioxidant activity that this cultivar exhibits. It should be emphasized that the most wanted biochemical, e.g., electrophoretical feature of identity, is the presence of lack of particular band/s on gel, in comparison of genotypes in detecting of polymorphism. Peaks height cannot be considered as identity feature: it indicates only quantitative, variable differences between cultivars. Similarly, Nishyama (2007) and other researches (Grozdanovic et al. 2014; Maddumage et al. 2013) have found intervarietal differences in actinidin activity on SDS-PAGE gel shown as differences of intensity of actinidin band. The difficulties in distinguishing A. arguta varieties on the basis of biochemical features of proteins can be explained by low range of genetic variability of materials on the market. We suppose that selection of new varieties was performed with a material of relatively limited pool of genetic variability. Very important factor which we could explain was the phenomena of low genetic variability. It is a common fact for this plant group where the used method of propagation is vegetative (non sexual procedure). It is known that the level of seed protein polymorphism, detected on gels, is usually lower than those of molecular markers, but it is suitable for distinguishing varieties, for the purposes of genotype identification. It aims for describing varietal identity and evaluation of genetic purity of seed samples and lots of agricultural crops, as cereals, grasses, and legume plants. We have demonstrated a possibility of successful extraction of proteins from singular seeds of kiwis A. arguta, followed by SDS-PAGE electrophoresis. Although obtained low intervarietal polymorphism of proteins was low and not allowed to distinguish all varieties of hardy kiwis, based on juice and seeds patterns, we supposed that proposed method can be suitable for distinguishing kiwi cultivars that were derived from more wide range of genetic pool. Our results are in accordance with others (Cooke 1989; Hameed et al. 2014; Shivashankar et al. 2010; Singh et al. 2011; Tamkoc and Arslan 2011) that SDS-PAGE method is the most suitable method of electrophoresis for the purpose of describing and distinguishing species and varieties of dicotyledonous plants-legumes, some vegetables-varieties such Chilli (Capsicum annuum L.), Brassica juncea genotypes, Poa L. (Poaceae) species, sunflower, tomato germplasm, and other dicots as pseudocereal plants. We had no problems with recognizing genetically different varieties as ‘Hayward’ and ‘Bidan’ cultivars, on the basis of method proposed by us. The problems with discrimination of more related varieties as A. arguta kiwis appeared. There is well documented data that seed protein markers generated by SDS-PAGE are suitable for discrimination of crop varieties (Cooke 1989), but some problems with distinguishing were described in the case of white and yellow lupines cultivars. Explanation was proposed that it may be caused by a limited genetic pool of genotypes for selected cultivars, and the similar problem has occurred in the case of examination of hardy kiwi cultivars. On the example of ‘Ananasnaya’ cultivar, we have confirmed genetic homogeneity of plants that are propagated by the way of vegetative process. It is known that on the market, new kiwi fruit genotypes and the number of seeds for sale increased. This factor implicates necessity of developing methods that can be suitable for laboratory check of identity material and evaluation of its genetic purity. Biochemical marker-protein pattern derived from single seeds can be used for plant breeder’s protection, before application of a new variety registration, also in the cases about doubts in samples identity on the market. The checking of crop seeds identity is based on the well-documented statement that biochemical and electrophoretical markers are independent of any environment changes, including year of harvest, and they are constant (Cooke 1989). Also others concluded that seed storage protein profiles could be useful markers in the studies of genetic diversity and genetic relationships of Poa species. SDS-PAGE method was preferred to inter- and intraspecific determinination of the genetic relationships of Poa species in order to facilitate genotype selection in breeding programs (Tamkoc and Arslan 2011). Our aim was to check whether this principle is obligated for kiwis too on the example of fruit proteins that were collected in 2 years. The answer for such question is positive. Any changes were not found in the electrophoregrams after the period of 3 weeks of fruits storage. Previously, other researchers have been interested in the problem whether any protein changes occur during maturing with ethylene treatment. After 2 days of maturing in 20 °C, only some minor changes in the intensity of 32 kDa band (by 1-D SDS-PAGE) were found (Park et al. 2008). One-week fruit maturing period caused about 1300 spots, which were recognized and corresponded to particular peptides. Nevertheless, the changes (presence of particular spots or lack of spots) were noticed only in the case of 32 spots (Li et al. 2014).
a The matrix of similarity of hardy kiwis and kiwi fruit cultivars. L1, ‘Bingo’; L2, ‘M1’; L3, ‘Ananasnaya’; L4, ‘Weiki’; L5, ‘Jumbo’; L6, ‘Geneva’; L7, ‘Hayward’; L8, ‘Bidan.’ Similarity coefficients of genetic similarity (GS) are shown at the matrix as decimal fractions. b UPGMA dendrogram of kiwi fruit cultivars based on protein data using genetic similarity (Nei and Li 1979) values in the matrix. 1, ‘Bingo’; 2, ‘M1’; 3, ‘Ananasnaya’; 4, ‘Weiki’; 5, ‘Jumbo’; 6, ‘Geneva’; 7, ‘Hayward’; 8, ‘Bidan’. 1–6—A. arguta, 7—A. deliciosa, 8—A. eriantha cultivars. Genetic similarity values are shown on the dendrogram in percentages, as whole numbers (GS × 100). c The densitometry profiles of proteins bands of kiwi fruit cultivars. 1, 'Bingo'; 2, ‘M1’; 3, ‘Ananasnaya’; 4, ‘Weiki’; 5, ‘Jumbo’ ; 6, ‘Geneva’; 7, ‘Hayward’; 8, ‘Bidan.’ 1–6—A. arguta, 7—A. deliciosa, 8—A. eriantha cultivars. The densitometry profiles of bands are shown as curves of peaks height. Height of peaks—maximum intensity on a range of 256 gray levels
During the process of 3-week period of storage at 1 °C and 90–95 % of RH in kiwi fruit harvested in commercial maturity, any possible changes of protein activity were not detected.
The significant progress in detecting kiwi fruit proteins was achieved by 2-D electrophoresis, followed by proteomics analysis. On the other hand, it is known that the use of 2-D electrophoresis, followed by proteomic analysis, was successful for solving problem of detecting biochemical changes during the storage of A. arguta fruits (Li et al. 2014). We supposed that the method used in our investigation (SDS-PAGE, followed by Coomassie staining) was not suitable for detecting minor changes due to low sensitivity of the stain used. On the other hand, the use of 2-D electrophoresis for kiwi genotypes, followed by proteome analysis, can be performed only for scientific work purposes, but not for routine tests—seed lots evaluation, tests performed by plant breeders, and seed testing laboratories. The present analysis of plants by protein markers as 1-D SDS-PAGE, IF, or A-PAGE markers represented the most effective testing of initial and breeding material (Cooke 1989). Our results are in line with Singh et al. (2011), where genetic variability among 59 B. juncea genotypes was assessed, based on morphological and biochemical markers. Based on the SDS-PAGE patterns, all genotypes were classified into five clusters.
Fourier Transform Infrared Spectra of Polyphenols
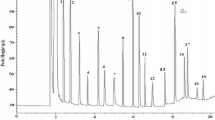
For illustration of the distinct patterns of the kiwi fruit cultivars, averaged spectra of each cultivar are given in Fig. 5. The IR spectra of kiwi A. arguta methanol extracts were compared between them (Fig. 5b, c) and also with standard kiwi fruit such as ‘Hayward’ and ‘Bidan’ in the range of common peaks (Fig. 5a). In our previous study, the IR spectra data showed that the main bands in the kiwi fruit samples slightly shifted (Park et al. 2015c). IR spectrum of polyphenols showed common peaks for all kiwi fruit samples at 3299–3300 cm−1, 2924–2930 cm−1, 1232–1235 cm−1, and 919–924 cm−1, except cultivar ‘M1’ (Fig. 5). A. arguta cultivars had bands of 3011–3007, 2851, 2353, and 2357 cm−1 (Fig. 5b, c). Then A. arguta cultivars showed peaks at 1743, 1737, 1651, 1603, 1605, 1317–1319, 1233, 1182, 1106–1140, 1042, 989, 924, 817, and 779 cm−1. ‘Hayward’ and ‘Bidan’ showed limited number of bands as A. arguta cultivars at 1716, 1603, 1593, 1402, 1340, 1232, 1027, 919, 866, 816, and 777 cm−1. A predominant band in the region of 1042–1140 cm−1 was assigned to the C-O stretching vibration of the sugars (Fig. 5). In all samples, the region at 3300–2900 cm−1 was dominated by one broad band, which represented the H-bonded O-H stretching of carbohydrate, carboxylic acids, and residual water. In the spectrum of the methanol soluble fraction of poly(rutin), a broad peak centered at 3300 cm−1 due to the vibration of O–H linkage of phenolic and hydroxyl groups and peaks at ca. 1 570 cm−1 ascribed to the C=C vibration of aromatic group were observed, the methanol-soluble and insoluble polymers. The doublets near 2925–2930 cm−1 were caused by the C-H asymmetric and symmetric stretching of the methyl C-H group. This may have been due to the presence of a phenolic hydroxyl group in the kiwi fruit polyphenol molecules. The characteristic functional groups stretches indicate the presence of alkane (3011 and 2930 cm−1), N-methyl (2851 cm−1), and C-O stretch (1182 cm−1) in the samples. The three bands between 1402 and 1603 cm−1 were attributed to aromatic ring vibrations. Multiple bands between 1140 and 1319 cm−1 comprised intricate absorption of the C-O stretch and C-O-H bending of phenols, carboxylic acids, and carbohydrates (Silverstein et al. 1981; Coates 2000; He et al. 2007; Lopez-Sánchez et al. 2010; Bureau et al. 2012). The spectral region giving the best results for phenolic compound prediction was 1010–1700 cm−1. This area corresponds to the spectral data of catechin obtained with the same ATR technique and showing important peaks between 1700 and 1000 cm−1. According to Coates (2000), this area shows phenolic compounds bands (C=C-C aromatic ring stretch, 1603 cm-1 in both A. arguta and A. deliciosa cultivars), to phenol OH bend (1402–1318 cm−1), to aromatic C-H in-plane bend (1232–966 cm−1), and to CO stretch of phenol (1232 cm−1). The total phenolic content showed numerous characteristic peaks in the spectral region (1010–1700 cm−1). Bands at around 1605, 1593, 1402, 1340, 1233, 1145, 1182, and 1042 cm−1 are found in all spectra with various relative intensities and minor, but varietal, characteristic shifting in absolute band positions. ‘M1’ differed from other A. arguta cultivars in the range of 1042–990 cm−1 and showed only one peak at 1015 cm−1. Our results are similar to others, showing the polyphenols differences in the spectral region giving the best results for phenolic compound prediction of 1010–1700 cm−1 (Bureau et al. 2012). In the region of 865–775 cm−1, ‘M1’ is similar to ‘Hayward’ and ‘Bidan.’ Thus, for each cultivar, a unique phenolic fingerprint in the IR spectral region could be obtained. Most remarkable are the differences from 1232 till 777 cm−1 (He et al. 2007; Lopez-Sánchez 2010; Bureau et al. 2012). A shift in the difference between the standard and the investigated samples can be explained by the extraction procedures of the total phenols. The matching (%) of the peaks in the region from 4000 to 700 cm−1 (short regions: 3400–2900 cm−1, 1800–900 cm−1, and 1100–700 cm−1) for Hayward/Bidan, Weiki/Bingo, and Jumbo/Ananasnaya was the highest (96–100 %). Cultivar ‘M1’ which showed differences in the electrophoretic separation as well showed lower matching in the same spectral region as: Hayward/M1, Ananasnaya/M1, Bidan/M1, Bingo/M1, and Geneva/M1 from 72 to 99 % (Fig. 5, Table 1). As it was discussed previously, by Bureau et al. (2012), the best matching region for polyphenols is 1100–700 cm−1 and the following values of matching were obtained for Hayward/M1 (90 %) and Bidan/M1 (93 %), and lower for Ananasnaya/M1 (84 %) and Geneva/M1 (76 %).
NMR Spectra of Polyphenols
NMR spectra of some cultivars are shown on Fig. 6. The assignment of 1H spectrum of kiwi fruit organic extract (DMSO) was obtained, where the main peaks appeared between 2 and 6 ppm. The spectra were similar for three cultivars with slight differences between 4–5 ppm for ‘Ananasnaya’ (Fig. 6c). In fact, the difference between the concentrations of the major and minor components of a metabolite mixture can be beyond the limited dynamic range of a NMR spectrometer. In this case, the limit of detection for the minor component in the mixture is higher than the limit of detection for the same compound isolated from the mixture. In the case of DMSO, mostly the sugars (Fig. 6a–c) were the main compounds and the aromatic part (Fig. 6d–f) was the minor (ppm): ‘Bidan’ (8.16, 7.22, 6.60, 6.23), ‘Hayward’ (8.17, 7.22, 6.64, 6.23), ‘Ananasnaya’ (8.15, 7.23, 6.58, 6.21). Using the NMR approach, it has been possible to identify primary as well as secondary metabolites of different fruits such as grape, orange, apple juice, mandarin, orange, kiwi fruits, mango, black raspberry, melon, watermelon, blueberry, and peaches (Sobolev et al. 2015; Capitani et al. 2010; Capitani et al. 2013a and 2013b). As it can be seen from the obtained data, all peaks in aromatic region in the cultivars were completely similar in DMSO solvent. It is known that phenolic compounds are poorly soluble in water. The mixture of water and methanol is usually used for extraction and subsequent NMR analysis (Fig. 6g, h): for ‘Hayward’ (9.37, 8.53, 8.34, 8.25, 8.16, 7.87, 7.80, 7.72, 7.46, 7.29, 7.23, 7.22, 7.16, 7.10, 6.98, 6.82, 6.78, 6.60, 6.46 ppm) and for ‘Ananasnaya’ (9.19, 8.05, 7.96, 7.95, 7.30, 7.10, 7.05, 6.92, 6.86, 6.73, 6.61, 6.42, 6.32 ppm). ‘Bidan’ showed exactly the same peaks as ‘Hayward.’ In ‘Ananasnaya,’ the number of peaks was sligthly lower that in the two varieties. In NMR-based analyses, the extraction procedure is probably the most critical step aimed to the quantitative transfer of the metabolites from the solid matrix into the solution. Metabolites identified in 1H-NMR spectra of kiwi fruit water extracts showed the following compounds: acids—ascorbic, citric, lactic, malic, quinic; sugars and sugar alcohols—β-Arabinose, α-d-fructofuranose, β-d-fructofuranose, β-d-fructopyranose, α-galactose, β-galactose, α-glucose, β-glucose, α-glucose-6P, β-glucose-6P, β-mannose, Myo-inositol, raffinose, sucrose, α-xylose, β-xylose; amino acids, peptides and derivatives—alanine, γ-amino-butyrate, arginine, asparagine, aspartate, glutamate, glutamine, histidine, isoleucine, leucine, lysine, phenylalanine, threonine, tryptophane, valine; alcohols, polyols, amines, aldehydes, ketones, esters—choline; nucleic acid derivatives—adenosine-triphosphate, uridine; aromatic compounds—(−)-epicatechin, neochlorogenic acid, quercetin-3-O-rhamnoside, O3-β-d-glucopyranosyltrans caffeic acid; O3-β-d-glucopyranosyl-cis-caffeic acid (Sobolev et al. 2015; Capitani et al. 2010; Capitani et al. 2013b). Sugars such as glucose, sucrose, and fructose have been detected in the investigated kiwi fruit cultivars. The observation of secondary metabolites in the 1H-NMR spectra of crude extracts can be hindered by the presence of major components (Sobolev et al. 2015; Capitani et al. 2010; Capitani et al. 2013b).
Conclusions
SDS-PAGE, which was used for the purpose of distinguishing hardy kiwi fruit varieties, was only partially successful due to low level of polymorphism between cultivars. Nevertheless, in the case of kiwi seed proteins, we have obtained a pattern consisted of up to 24 bands. We expected that after improving electrophoresis protocols this feature can be suitable both for distinguishing hardy varieties and for kiwi genotypes that are derived from plant genetic sources with more wide spectrum of genetic variability. Our results, especially about a genetic homogeneity, between singular seeds can be considered as having a big value for kiwi fruits as a representative of plants that are propagated not only by sexual way but also by vegetative reproduction. We can propose a new tool of checking of vegetative reproduction which was properly done. The matching results for the first time showed that IR and NMR spectra can be used for a rapid estimation of extracted bioactive compounds, as an additional indication of similarity and differences between the cultivars. SDS-PAGE protein separation gave better estimation between the kiwi fruit cultivars than the extracted polyphenols and their measurements by FT-IR and NMR spectra.
References
Afshar-Mohammadian M, Rahimi-Koldeh J, Sajedi RH (2011) The comparison of protease activity and total protein in three cultivars of kiwifruit of Northern Iran during fruit development. Acta Physiol Plant 33:343–348
Anastasiadi M, Zira A, Magiatis P, Haroutounian SA, Skaltsounis AL, Mikros E (2009) 1H NMR-based metabonomics for the classification of Greek wines according to variety, region, and vintage. Comparison with HPLC data. J Agric Food Chem 57:11067–11074
Bureau S, Ścibisz I, Le Bourvellec C, Renard CMGC (2012) Effect of sample preparation on the measurement of sugars, organic acids, and polyphenols in apple fruit by mid infrared spectroscopy. J Agric Food Chem 60:3551–3563
Capitani D, Mannina L, Proietti N, Sobolev AP, Tomassini A, Miccheli A, di Cocco ME, Capuani G, de Salvador R, Delfini M (2010) Monitoring of metabolic profiling and water status of Hayward kiwifruits by nuclear magnetic resonance. Talanta 82:1826–1838
Capitani D, Sobolev AP, Tomassini A, Sciubba F, de Salvador FR, Mannina L, Delfini M (2013a) Peach fruit: metabolic comparative analysis of two varieties with different resistances to insect attacks by NMR spectroscopy. J Agric Food Chem 61:1718–1726
Capitani D, Mannina L, Proietti N, Sobolev AP, Tomassini A, Miccheli A, Di Cocco ME, Capuani G, de Salvador FR, Delfini M (2013b) Metabolic profiling and outer pericarp water state in Zespri, CI.GI and Hayward kiwifruits. J Agric Food Chem 61:1727–1740
Ciardello M, Tamburrini M, Tuppo L, Carratore V, Giovane A, Mattei B, Camardella L (2004) Pectin methylesterase from kiwi and kaki fruits: purification, characterization, and role of pH in the enzyme regulation and interaction with the kiwi proteinaceous inhibitor. J Agric Food Chem 52:7700–7703
Coates J (2000) Interpretation of infrared spectra, a practical approach. In: Meyers RA (ed) Encyclopedia of analytical chemistry. Wiley, Chichester, pp 10815–10837
Cooke R (1989) Electrophoresis in plant testing and breeding. Electrophoresis 2:171–261
Grozdanovic MM, Ostojic S, Aleksic I, Andjelkovic U, Petersen A, Gavrovic-Jankulovic M (2014) Active actinidin retains function upon gastro-intestinal digestion and is more thermostable than the E-64-inhibited counterpart. J Sci Food Agric 94:3046–3052
Hameed A, Gul A, Gulzar T (2014) Characterization of tomato germplasm through seed storage protein profiling by SDS-PAGE. Pak J Bot 46:827–832
He J, Rodriguez-Saona LE, Giusti MM (2007) Midinfrared spectroscopy for juice authentications rapid differentiation of commercial juices. J Agric Food Chem 55:4443–4452
Huang H, Gong J-J, Wang S-M, He Z-C, Zhang Z-H, Li J-Q (2000) Genetic diversity in the genus Actinidia. Chin Biodivers 8:1–12
Huang H, Li Z, Li J, Kubisiak TL, Layne DR (2002) Phyllogenetic relationships in Actinidia as revealed by RAPD analysis. J Am Sci 127:759–766
Jo YS, Cho HS, Park MY, Bang GP (2007) Selection of a sweet Actinidia eriantha, ‘Bidan’. VI International Symposium on Kiwifruit ISHS. Acta Horticult 753:253–257
Laemmli UK (1970) Cleavage of structural proteins the assembly of bacteriophage T4. Nature 227:680–685
Latocha P, Krupa T (2007) Morphological, chemical and sensory analyses of promising genotypes of hardy kiwifruit (Actinidia Lindl.) obtained in the breeding programme at WULS. Ann Warsaw Univ of Life Sci-SGGW, Hortic Landsc Archit 28:111–119
Latocha P, Olszewska-Kaczyńska I (2003) Preliminary morphological, chemical and sensory analysis of fruit of different Actinidia genotypes (Actinidia Lindl.). Ann Warsaw Univ Life Sci-SGGW, Hortic Landsc Archit 24:91–104
Li S-Q, Xin G, Zhang B, Liu C-J (2014) Proteome change of hardy kiwifruit Turing softening. J Chem Pharm Res 6:2208–2212
López-Sánchez M, Ayora-Cañada MJ, Molina-Díaz A (2010) Olive fruit growth and ripening as seen by vibrational spectroscopy. J Agric Food Chem 58:82–87
Maddumage R, Nieuwenhuizen N, Bulley SM, Cooney JM, Green SA, Atkinson G (2013) Diversity and relative levels of actinidin, kiwellin, and thaumatin-like allergensin 15 varieties of kiwifruit (Actinidia). J Agric Food Chem 61:728–739
Miraghaee SS, Mostafaie A, Kiani S, Kahrizi D (2011) Investigation on protein pattern in kiwifruit (Actinidia deliciosa). World Appl Sci 15:1398–1402
Miraghaee SS, Mostafaie A, Kahrizi D (2012) Acclimatization related proteins and factors in somaclone lines in kiwifruit (Actinidia deliciosa). J Food Agric Environ 10:198–202
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A 76:5269–5373
Nishyama I (2007) Fruits of the Actinidia genus. Adv Food Nutr Res 52:293–324
Park Y-S, Jung S-T, Kang S-G, Heo B-G, Arancibia-Avilla P, Toledo F, Drzewiecki J, Namiesnik J, Gorinstein S (2008) Antioxidants and proteins in ethylene-treated kiwifruits. Food Chem 107:640–648
Park Y-S, Ham K-S, Kang S-G, Park Y-K, Namiesnik J, Leontowicz H, Leontowicz M, Ezra A, Trakhtenberg S, Gorinstein S (2012a) Organic and conventional kiwifruit, myths versus reality: antioxidant, antiproliferative, and health effects. J Agric Food Chem 60:6984–6993
Park Y-S, Heo B-G, Ham K-S, Kang S-G, Park Y- K, Nemirovsky A, Tashma Z, Gorinstein S, Leontowicz H, Leontowicz M (2012b) Analytical determination of bioactive compounds as an indication of fruit quality. J AOAC Intern 95:1725–1732
Park Y-S, Im M-H, Ham K-S, Kang S-G, Park Y-K, Namiesnik J, Leontowicz H, Leontowicz M, Katrich E, Gorinstein S (2013) Nutritional and pharmaceutical properties in bioactive compounds in organic and conventional growing kiwifruit. Plant Foods Hum Nutr 68:57–64
Park Y-S, Im MH, Ham K-S, Kang S-G, Park Y-K, Namiesnik J, Leontowicz H, Leontowicz M, Trakhtenberg S, Gorinstein S (2015a) Quantitative assessment of the main antioxidant compounds, antioxidant activities and FTIR spectra from commonly consumed fruits, compared to standard kiwi fruit. LWT - Food Sci Technol 63:346–352
Park Y-S, Polovka M, Martinez-Ayala AL, Gonzáles-Aguilar GA, Ham K-S, Kang S-G, Park Y-K, Heo B-G, Namiesnik J, Gorinstein S (2015b) Fluorescence studies by quenching and protein unfolding on the interaction of bioactive compounds in water extracts with human serum albumin. J Lumin 160:71–77
Park Y-S, Leontowicz M, Leontowicz H, Ham K-S, Kang S-G, Park Y-K, Rombola AD, Katrich E, Gorinstein S (2015c) Fluorescence and ultraviolet spectroscopic evaluation of phenolic compounds, antioxidant and binding activities in some kiwi cultivars. Spectrosc Lett 48:586–592
Shivashankar S, Thimmareddy C, Roy TK (2010) Polygalacturonase inhibitor protein from fruits of anthracnose resistant and susceptible varieties of Chilli (Capsicum annuum L.). Indian J Biochem Biophys 47:243–248
Silverstein RM, Bassler GC, Morrill TC (1981) Spectrometric identification of organic compounds. Wiley, Chichester
Singh VK, Bangari G, Singh D, Tewari S (2011) Evaluation of genetic variation within Brassica juncea genotypes using biochemical and morphological markers. Vegetos 24:208–214
Sobolev AP, Mannina L, Proietti N, Carradori S, Daglia M, Giusti AM, Antiochia R, Capitani D (2015) Untargeted NMR-based methodology in the study of fruit metabolites. Molecules 20:4088–4108
Tamburrini M, Cerasuolo I, Carratore V, Stanziola AA, Zofra S, Romano L, Camardella L, Ciardiello MA (2005) Kiwellin, a novel protein from kiwi fruit. Purification, biochemical characterisation and identification as an allergen. Protein J 24:423–429
Tamkoc A, Arslan E (2011) Inter and intra-specific variation in SDS-PAGE of seed proteins of three Poa L. (Poaceae) species. Pak J Bot 43:1105–1110
Testolin R, Ferguson AR (1997) Isozyme polymorphism in the genus Actinidia and the origin of the kiwifruit genome. Syst Bot 22:685–700
Tupo L, Gangrieco I, Palazzo P, Bernardi ML, Scala E, Carratore V, Tamburrini M, Mari A, Ciardiello MA (2008) Kiwellin, a modular protein from green and gold kiwi fruits: evidence of in vivo and in vitro processing and IgE binding. J Agric Food Chem 56:3812–3817
Vergara-Barberan M, Lerma-Garcia MJ, Herrero-Martinez JM, Simo-Alfonso EF (2015) Use of an enzyme-assisted method to improve protein extraction from olive leaves. Food Chem 169:28–33
Zhu DY, Lawes GS (1990) Characterization of kiwifruit (Actinidia deliciosa) cultivars by gel electrophoresis. Acta Hortic 282:395–398
Acknowledgments
The authors are thankful to Dr. Elena Katrich (School of Pharmacy, Hebrew University of Jerusalem) for her technical assistance in FT-IR spectra.
Funding
This study was partially funded by a grant from National Science Center 2012/05/B/NZ9/03327, Poland.
Conflict of Interest
All authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Drzewiecki, J., Latocha, P., Leontowicz, H. et al. Analytical Methods Applied to Characterization of Actinidia arguta, Actinidia deliciosa, and Actinidia eriantha Kiwi Fruit Cultivars. Food Anal. Methods 9, 1353–1366 (2016). https://doi.org/10.1007/s12161-015-0309-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-015-0309-1