Abstract
Background
Long noncoding RNA (lncRNAs) GMDS-AS1 has been reported as a tumor regulator in tumor growth and metastasis, but its effect in hepatocellular carcinoma (HCC) remains unclear. ESET, a histone H3K9 methyl-transferase, is involved in epigenomic regulation of tumor progression in multiple cancers. However, the correlation between ESET and lncRNA in HCC is less reported.
Methods
Quantitative real-time PCR (qRT-PCR) was taken to determine the expression of ESET and GMDS-AS1. Western blot was taken to determine the target protein levels of ESET and GMDS-AS1. Online database and bioinformatics analysis were used to screen abnormally expressed genes. Luciferase assay was performed to confirm the binding of GMDS-AS1 and PSMB1. Ki67 and Edu were used for evaluated the proliferation of tumor cells. ChIP assay was performed to verify the relationship between H3K9me1 and lncRNA GMDS-AS1 promoter. Transwell was taken to determine the migration and invasion ability of tumor cells. CCK-8 was used for determining the viability of tumor cells. Flow cytometry was performed to detect the cell cycle of tumor cells.
Results
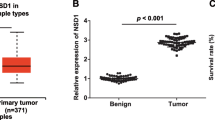
The expression of GMDS-AS1 was decreased and the expression of ESET was increased in HCC. GMDS-AS1 inhibition contributed to tumor development, and this effect was closely related to epigenetic inhibition of GMDS-AS1 by ESET. PSMB1, a downstream target of GMDS-AS1, promoted the tumor proliferation and was negatively regulated by GMDS-AS1.
Conclusion
Our result demonstrates anti-tumorigenic traits of lncRNA GMDS-AS1 in HCC and explains its pattern of regulation mediated by ESET. Our work unmasked an essential role of GMDS-AS1 in HCC progression and detected a novel pathway for ESET to promote HCC.






Similar content being viewed by others
Data availability
The sequencing data used in Fig. 4A, B were download from the GEO data base [https://www.ncbi.nlm.nih.gov/geo/, GSE138178]. The other data are available in the method of this article.
References
Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–62. https://doi.org/10.1056/NEJMra1713263.
Li HM, Ye ZH. Microenvironment of liver regeneration in liver cancer. Chin J Integr Med. 2017;23(7):555–60. https://doi.org/10.1007/s11655-017-2806-0.
Li HM. Microcirculation of liver cancer, microenvironment of liver regeneration, and the strategy of Chinese medicine. Chin J Integr Med. 2016;22(3):163–7. https://doi.org/10.1007/s11655-016-2460-y.
Bruix J, Han KH, Gores G, Llovet JM, Mazzaferro V. Liver cancer: approaching a personalized care. J Hepatol. 2015;62(1 Suppl):S144–56. https://doi.org/10.1016/j.jhep.2015.02.007.
Xia H, Hui KM. Emergence of aspirin as a promising chemopreventive and chemotherapeutic agent for liver cancer. Cell Death Dis. 2017;8(10): e3112. https://doi.org/10.1038/cddis.2017.513.
Buonaguro L, HEPAVAC Consortium. New vaccination strategies in liver cancer. Cytokine Growth Factor Rev. 2017;36:125–9. https://doi.org/10.1016/j.cytogfr.2017.06.010.
Stillman B. Histone modifications: insights into their influence on gene expression. Cell. 2018;175(1):6–9. https://doi.org/10.1016/j.cell.2018.08.032.
Lomvardas S, Maniatis T. Histone and DNA modifications as regulators of neuronal development and function. Cold Spring Harb Perspect Biol. 2016. https://doi.org/10.1101/cshperspect.a024208.
Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32(1):42–56. https://doi.org/10.1016/j.tig.2015.10.007.
Yan J, Chen SA, Local A, Liu T, Qiu Y, Dorighi KM, et al. Histone H3 lysine 4 monomethylation modulates long-range chromatin interactions at enhancers. Cell Res. 2018;28(2):204–20. https://doi.org/10.1038/cr.2018.1.
Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48(4):491–507. https://doi.org/10.1016/j.molcel.2012.11.006.
Padeken J, Methot SP, Gasser SM. Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance. Nat Rev Mol Cell Biol. 2022;23(9):623–40. https://doi.org/10.1038/s41580-022-00483-w.
Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3–K9 methyltransferase ESET is essential for early development. Mol Cell Biol. 2004;24(6):2478–86. https://doi.org/10.1128/MCB.24.6.2478-2486.2004.
Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell. 2003;12(2):475–87. https://doi.org/10.1016/j.molcel.2003.08.007.
Lazaro-Camp VJ, Salari K, Meng X, Yang S. SETDB1 in cancer: overexpression and its therapeutic implications. Am J Cancer Res. 2021;11(5):1803–27.
Wong CM, Wei L, Law CT, Ho DW, Tsang FH, Au SL, et al. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology. 2016;63(2):474–87. https://doi.org/10.1002/hep.28304.
Fei Q, Shang K, Zhang J, Chuai S, Kong D, Zhou T, et al. Histone methyltransferase SETDB1 regulates liver cancer cell growth through methylation of p53. Nat Commun. 2015;6:8651. https://doi.org/10.1038/ncomms9651.
Zhang Y, Huang J, Li Q, Chen K, Liang Y, Zhan Z, et al. Histone methyltransferase SETDB1 promotes cells proliferation and migration by interacting withTiam1 in hepatocellular carcinoma. BMC Cancer. 2018;18(1):539. https://doi.org/10.1186/s12885-018-4464-9.
Jathar S, Kumar V, Srivastava J, Tripathi V. Technological developments in lncRNA biology. Adv Exp Med Biol. 2017;1008:283–323. https://doi.org/10.1007/978-981-10-5203-3_10.
Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. https://doi.org/10.1007/978-981-10-5203-3_1.
Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang C, et al. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer. 2017;16(1):123. https://doi.org/10.1186/s12943-017-0702-z.
Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration EMT and metastasis. Int J Cancer. 2016;139(2):269–80. https://doi.org/10.1002/ijc.30039.
Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17(1):106–16. https://doi.org/10.1093/bib/bbv031.
Zhao M, Xin XF, Zhang JY, Dai W, Lv TF, Song Y. LncRNA GMDS-AS1 inhibits lung adenocarcinoma development by regulating miR-96-5p/CYLD signaling. Cancer Med. 2020;9(3):1196–208. https://doi.org/10.1002/cam4.2776.
Rousseau A, Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nat Rev Mol Cell Biol. 2018;19(11):697–712. https://doi.org/10.1038/s41580-018-0040-z.
Catalgol B. Proteasome and cancer. Prog Mol Biol Transl Sci. 2012;109:277–93. https://doi.org/10.1016/B978-0-12-397863-9.00008-0.
Yuan F, Ma Y, You P, Lin W, Lu H, Yu Y, et al. A novel role of proteasomal beta1 subunit in tumorigenesis. 2013. Biosci Rep. https://doi.org/10.1042/BSR20130013.
Rimassa L, Pressiani T, Merle P. Systemic treatment options in hepatocellular carcinoma. Liver Cancer. 2019;8(6):427–46. https://doi.org/10.1159/000499765.
Bouattour M, Mehta N, He AR, Cohen EI, Nault JC. Systemic treatment for advanced hepatocellular carcinoma. Liver Cancer. 2019;8(5):341–58. https://doi.org/10.1159/000496439.
Keane FK, Wo JY, Zhu AX, Hong TS. Liver-Directed radiotherapy for hepatocellular carcinoma. Liver Cancer. 2016;5(3):198–209. https://doi.org/10.1159/000367764.
Han TS, Ban HS, Hur K, Cho HS. The epigenetic regulation of HCC metastasis. Int J Mol Sci. 2018. https://doi.org/10.3390/ijms19123978.
Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. https://doi.org/10.1126/science.1063127.
Han J, Qu H, Han M, Ding Y, Xie M, Hu J, et al. MSC-induced lncRNA AGAP2-AS1 promotes stemness and trastuzumab resistance through regulating CPT1 expression and fatty acid oxidation in breast cancer. Oncogene. 2021;40(4):833–47. https://doi.org/10.1038/s41388-020-01574-8.
Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–8. https://doi.org/10.1136/jmedgenet-2015-103334.
Bi G, Liang J, Zhao M, Zhang H, Jin X, Lu T, et al. miR-6077 promotes cisplatin/pemetrexed resistance in lung adenocarcinoma via CDKN1A/cell cycle arrest and KEAP1/ferroptosis pathways. Mol Ther Nucleic Acids. 2022;28:366–86. https://doi.org/10.1016/j.omtn.2022.03.020.
Fricker LD. Proteasome inhibitor drugs. Annu Rev Pharmacol Toxicol. 2020;60:457–76. https://doi.org/10.1146/annurev-pharmtox-010919-023603.
Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A. Structure and function of the 26S proteasome. Annu Rev Biochem. 2018;87:697–724. https://doi.org/10.1146/annurev-biochem-062917-011931.
Acknowledgements
We greatly appreciate the support from the Guangxi University young and middle-aged teachers research ability enhancement project (KY2016LX257), Guangxi University Key Laboratory Project (kfkt2016009) and High-level talents research project of the Affiliated Hospital of Youjiang Medical University for Nationalities (Y20196303).
Author information
Authors and Affiliations
Contributions
JH: Conceptualization, Writing—Original Draft, Writing–Editing; TZ: Validation, Supervision, Funding acquisition, Project administration; GL: Methodology, Investigation, Resources, Experimentation; SW: Experimentation; RQ: Formal analysis, Data Curation, Visualization.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval and consent to participate
This study was approved by the Ethics Committee of the Affiliated Hospital of Youjiang Medical University for Nationalities. The study was conducted in accordance with the Declaration of Helsinki.
Consent for publication
All authors have read and approved the final manuscript.
Human participants and/or animals and Informed consent
This study involves animal experiments were approved by the Ethics Committee of the Affiliated Hospital of Youjiang Medical University for Nationalities. This study does not involve human participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, J., Zhong, T., Li, G. et al. Epigenetic inhibition of lncRNA GMDS-AS1 by methyltransferase ESET promoted cell viability and metastasis of hepatocellular carcinoma. Clin Transl Oncol 25, 1793–1804 (2023). https://doi.org/10.1007/s12094-023-03077-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03077-w