Abstract
Purpose
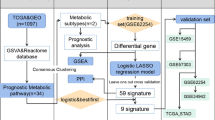
The growth and aggressiveness of Stomach adenocarcinoma (STAD) is significantly affected by basic metabolic changes. This study aimed to identify metabolic gene prognostic signatures in STAD.
Methods
An integrative analysis of datasets from the Cancer Genome Atlas and Gene Expression Omnibus was performed. A metabolic gene prognostic signature was developed using univariable Cox regression and Kaplan–Meier survival analysis. A nomogram model was developed to predict the prognosis of STAD patients. Finally, Gene Set Enrichment Analysis (GESA) was used to explore the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways significantly associated with the risk grouping.
Results
A total of 327 metabolism-related differentially expressed genes were identified. Three subtypes of STAD were identified and nine immune cell types, including memory B cell, resting and activated CD4+ memory T cells, were significantly different among the three subgroups. A risk score model including nine survival-related genes which could separate high-risk patients from low-risk patients was developed. The prognosis of STAD patients likely benefited from lower expression levels of genes, including ABCG4, ABCA6, GPX8, KYNU, ST8SIA5, and CYP19A1. Age, radiation therapy, tumor recurrence, and risk score model status were found to be independent risk factors for STAD and were used for developing a nomogram. Nine KEGG pathways, including spliceosome, pentose phosphate pathway, and citrate TCA cycle were significantly enriched in GESA.
Conclusion
We propose a metabolic gene signature and a nomogram for STAD which might be used for predicting the survival of STAD patients and exploring prognostic markers.









Similar content being viewed by others
References
Ramazani Y, Mardani E, Najafi F, Moradinazar M, Amini M. Epidemiology of gastric cancer in North Africa and the middle east from 1990 to 2017. J Gastrointest Cancer. 2021;52:1046–53.
Yang L, Ying X, Liu S, Lyu G, Xu Z, Zhang X, Li H, Li Q, Wang N, Ji J. Gastric cancer: epidemiology, risk factors and prevention strategies. Chin J Cancer Res. 2020;32:695–704.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49.
Okugawa Y, Mohri Y, Tanaka K, Kawamura M, Saigusa S, Toiyama Y, Ohi M, Inoue Y, Miki C, Kusunoki M. Metastasis-associated protein is a predictive biomarker for metastasis and recurrence in gastric cancer. Oncol Rep. 2016;36:1893–900.
Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95.
Tabe Y, Lorenzi PL, Konopleva M. Amino acid metabolism in hematologic malignancies and the era of targeted therapy. Blood. 2019;134:1014–23.
D’Aniello C, Patriarca EJ, Phang JM, Minchiotti G. Proline Metabolism in Tumor Growth and Metastatic Progression. Front Oncol. 2020;10:776.
Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci. 2016;73:377–92.
Glunde K, Jacobs MA, Bhujwalla ZM. Choline metabolism in cancer: implications for diagnosis and therapy. Expert Rev Mol Diagn. 2006;6:821–9.
Dai M, Ma T, Niu Y, Zhang M, Zhu Z, Wang S, Liu H. Analysis of low-molecular-weight metabolites in stomach cancer cells by a simplified and inexpensive GC/MS metabolomics method. Anal Bioanal Chem. 2020;412:2981–91.
Lario S, Ramírez-Lázaro MJ, Sanjuan-Herráez D, Brunet-Vega A, Pericay C, Gombau L, Junquera F, Quintás G, Calvet X. Plasma sample based analysis of gastric cancer progression using targeted metabolomics. Sci Rep. 2017;7:17774.
Chen JL, Tang HQ, Hu JD, Fan J, Hong J, Gu JZ. Metabolomics of gastric cancer metastasis detected by gas chromatography and mass spectrometry. World J Gastroenterol. 2010;16:5874–80.
Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–10.
Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–56.
Oh SC, Sohn BH, Cheong JH, Kim SB, Lee JE, Park KC, Lee SH, Park JL, Park YY, Lee HS, et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat Commun. 2018;9:1777.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:47.
Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–8.
Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng Z, Zhu G, Qi J, Ma H, Nian H, et al. RNA-seq analyses of multiple meristems of soybean: novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol. 2014;14:169.
Huo Y, Li S, Liu J, Li X, Luo XJ. Functional genomics reveal gene regulatory mechanisms underlying schizophrenia risk. Nat Commun. 2019;10:670.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
Zhang X, Ren L, Yan X, Shan Y, Liu L, Zhou J, Kuang Q, Li M, Long H, Lai W. Identification of immune-related lncRNAs in periodontitis reveals regulation network of gene-lncRNA-pathway-immunocyte. Int Immunopharmacol. 2020;84:106600.
Wang P, Wang Y, Hang B, Zou X, Mao JH. A novel gene expression-based prognostic scoring system to predict survival in gastric cancer. Oncotarget. 2016;7:55343–51.
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–59.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7.
Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385–95.
Goeman JJ. L1 penalized estimation in the Cox proportional hazards model. Biom J. 2010;52:70–84.
Harrell FE Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–87.
Mayr A, Schmid M. Boosting the concordance index for survival data–a unified framework to derive and evaluate biomarker combinations. PLoS ONE. 2014;9:e84483.
Shan S, Chen W, Jia JD. Transcriptome analysis revealed a highly connected gene module associated with cirrhosis to hepatocellular carcinoma development. Front Genet. 2019;10:305.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–91.
Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu Rev Nutr. 2008;28:73–95.
Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3.
Gong PJ, Shao YC, Huang SR, Zeng YF, Yuan XN, Xu JJ, Yin WN, Wei L, Zhang JW. Hypoxia-associated prognostic markers and competing endogenous RNA co-expression networks in breast cancer. Front Oncol. 2020;10:579868.
Du Y, Ji Z, Liao J, Liu H, Peng H. Identification of molecular subtypes in head and neck squamous cell carcinoma based on dysregulated immune LncRNAs. J Oncol. 2022;2022:9702789.
Li G, Wu Z, Gu J, Zhu Y, Zhang T, Wang F, Huang K, Gu C, Xu K, Zhan R, et al. Metabolic signature-based subtypes may pave novel ways for low-grade glioma prognosis and therapy. Front Cell Dev Biol. 2021;9:755776.
Shen K, Liu T. Comprehensive analysis of the prognostic value and immune function of immune checkpoints in stomach adenocarcinoma. Int J Gen Med. 2021;14:5807–24.
Derks S, de Klerk LK, Xu X, Fleitas T, Liu KX, Liu Y, Dietlein F, Margolis C, Chiaravalli AM, Da Silva AC, et al. Characterizing diversity in the tumor-immune microenvironment of distinct subclasses of gastroesophageal adenocarcinomas. Ann Oncol. 2020;31:1011–20.
Wang X, Hu LP, Qin WT, Yang Q, Chen DY, Li Q, Zhou KX, Huang PQ, Xu CJ, Li J, et al. Identification of a subset of immunosuppressive P2RX1-negative neutrophils in pancreatic cancer liver metastasis. Nat Commun. 2021;12:174.
Tomas L, Edsfeldt A, Mollet IG, Perisic Matic L, Prehn C, Adamski J, Paulsson-Berne G, Hedin U, Nilsson J, Bengtsson E, et al. Altered metabolism distinguishes high-risk from stable carotid atherosclerotic plaques. Eur Heart J. 2018;39:2301–10.
Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, Hess JM, Uzunangelov V, Walter V, Danilova L, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:204-20.e15.
Jia Y, Dai J, Zeng Z. Potential relationship between the selenoproteome and cancer. Mol Clin Oncol. 2020;13:83.
Huang R, Zheng Z, Xian S, Zhang J, Jia J, Song D, Yan P, Yin H, Hu P, Zhu X, et al. Identification of prognostic and bone metastatic alternative splicing signatures in bladder cancer. Bioengineered. 2021;12:5289–304.
Wu S, Dai X, Xie D. Identification and validation of an immune-related RNA signature to predict survival of patients with head and neck squamous cell carcinoma. Front Genet. 2019;10:1252.
Batcioglu K, Mehmet N, Ozturk IC, Yilmaz M, Aydogdu N, Erguvan R, Uyumlu B, Genc M, Karagozler AA. Lipid peroxidation and antioxidant status in stomach cancer. Cancer Invest. 2006;24:18–21.
Hong C, Yang S, Wang Q, Zhang S, Wu W, Chen J, Zhong D, Li M, Li L, Li J, et al. Epigenetic age acceleration of stomach adenocarcinoma associated with tumor stemness features, immunoactivation, and favorable prognosis. Front Genet. 2021;12:563051.
Hazard L, O’Connor J, Scaife C. Role of radiation therapy in gastric adenocarcinoma. World J Gastroenterol. 2006;12:1511–20.
Duan S, Wang P, Liu F, Huang H, An W, Pan S, Wang X. Novel immune-risk score of gastric cancer: a molecular prediction model combining the value of immune-risk status and chemosensitivity. Cancer Med. 2019;8:2675–85.
Polterauer S, Grimm C, Hofstetter G, Concin N, Natter C, Sturdza A, Pötter R, Marth C, Reinthaller A, Heinze G. Nomogram prediction for overall survival of patients diagnosed with cervical cancer. Br J Cancer. 2012;107:918–24.
Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015;16:e173–80.
Funding
This work was supported by Changzhou Sci&Tech (CJ20200097) and Young Science & Technology Project of Changzhou Health Commission (QN202033).
Author information
Authors and Affiliations
Contributions
YG and SW conceived and designed the study, and drafted the manuscript. SD, SC and GC collected, analyzed and interpreted the experimental data. KB, HY and YJ revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by Ethical Committee of The Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University and conducted in accordance with the ethical standards.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Informed consent
Yes.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gong, Y., Wu, S., Dong, S. et al. Development of a prognostic metabolic signature in stomach adenocarcinoma. Clin Transl Oncol 24, 1615–1630 (2022). https://doi.org/10.1007/s12094-022-02809-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-022-02809-8