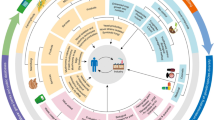
Abstract
Bioactive molecules of microbial origin are finding increasing biotechnological applications. Their sources range from the terrestrial, marine, and endophytic to the human microbiome. These biomolecules have unique chemical structures and related groups, which enable them to improve the efficiency of the bioprocesses. This review focuses on the applications of biomolecules in bioremediation, agriculture, food, pharmaceutical industries, and human health.
Similar content being viewed by others
References
Scherlach K, Hertweck C (2021) Mining and unearthing hidden biosynthetic potential. Nat Commun 12:3864. https://doi.org/10.1038/s41467-021-24133-5
Matuszewska A, Jaszek M, Stefaniuk D et al (2018) Anticancer, antioxidant, and antibacterial activities of low molecular weight bioactive subfractions isolated from cultures of wood degrading fungus Cerrena unicolor. PLoS ONE 13:e0197044. https://doi.org/10.1371/journal.pone.0197044
Adeleke BS, Babalola OO (2021) Biotechnological overview of agriculturally important endophytic fungi. Hortic Environ Biotechnol 62:507–520. https://doi.org/10.1007/s13580-021-00334-1
Abdel-Mageed W, Milne B, Wagner M et al (2010) Dermacozines, a new phenazine family from deep-sea dermacocci isolated from a Mariana Trench sediment. Org Biomol Chem 8:2352–2362. https://doi.org/10.1039/c001445a
Graça A, Bondoso J, Gaspar H et al (2013) Antimicrobial activity of heterotrophic bacterial communities from the marine sponge Erylus discophorus (astrophorida, geodiidae). PLoS ONE 8:e78992. https://doi.org/10.1371/journal.pone.0078992
Kalia VC (2017) Mining metagenomes for novel bioactive molecules. In: Kalia VC, Shouche Y, Purohit HJ, Rahi P (eds) Mining of microbial wealth and metagenomics. Springer, Singapore, pp 1–9. https://doi.org/10.1007/978-981-10-5708-3_1
Kalia VC (2017) The dawn of the era of bioactive compounds. In: Kalia VC (ed) Metabolic engineering for bioactive compounds. Springer, Singapore, pp 3–10. https://doi.org/10.1007/978-981-10-5511-9_1
Saini AK, Kalia VC (2017) Potential challenges and alternative approaches in metabolic engineering of bioactive compounds in industrial set up. In: Saini AK, Kalia VC (eds) Metabolic engineering for bioactive compounds. Springer, Singapore, pp 405–412. https://doi.org/10.1007/978-981-10-5511-9_19
Radivojevic J, Skaro S, Senerovic L et al (2016) Polyhydroxyalkanoate-based 3-hydroxyoctanoic acid and its derivatives as a platform of bioactive compounds. Appl Microbiol Biotechnol 100:161–172. https://doi.org/10.1007/s00253-015-6984-4
Mazzoli R, Riedel K, Pessione E (2017) Bioactive compounds from microbes. Front Microbiol 8:392. https://doi.org/10.3389/fmicb.2017.00392
Elfeki M, Alanjary M, Green SJ et al (2018) Assessing the efficiency of cultivation techniques to recover natural product biosynthetic gene populations from sediment. ACS Chem Biol 13:2074–2081. https://doi.org/10.1021/acschembio.8b00254
O’Mahony MM, Henneberger R, Selvin J et al (2015) Inhibition of the growth of Bacillus subtilis DSM10 by a newly discovered antibacterial protein from the soil metagenome. Bioengineered 6:89–98. https://doi.org/10.1080/21655979.2015.1018493
Jin Z, Di Rienzi SC, Janzon A et al (2015) Novel rhizosphere soil alleles for the enzyme 1-aminocyclopropane-1-carboxylate deaminase queried for function with an in vivo competition assay. Appl Environ Microbiol 82:1050–1059. https://doi.org/10.1128/AEM.03074-15
Luo W, Xu Z, Riber L et al (2016) Diverse gene functions in a soil mobilome. Soil Biol Biochem 101:175–183. https://doi.org/10.1016/j.soilbio.2016.07.018
Meneses C, Silva B, Medeiros B et al (2016) A metagenomic advance for the cloning and characterization of a cellulase from red rice crop residues. Molecules 21:e831. https://doi.org/10.3390/molecules21070831
Ilmberger N, Streit WR (2017) Screening for cellulase encoding clones in metagenomic libraries. Methods Mol Biol 1539:205–217. https://doi.org/10.1007/978-1-4939-6691-2_12
Deng J, Gao H, Gao Z et al (2017) Identification and molecular characterization of a metagenome-derived L-lysine decarboxylase gene from subtropical soil microorganisms. PLoS ONE 12:e0185060. https://doi.org/10.1371/journal.pone.0185060
Huang Y, Huang Y, Ji X et al (2021) Green chemical and biological synthesis of cadaverine: recent development and challenges. RSC Adv 11:23922–23942. https://doi.org/10.1039/d1ra02764f
Bao YJ, Xu Z, Li Y et al (2017) High-throughput metagenomic analysis of petroleum-contaminated soil microbiome reveals the versatility in xenobiotic aromatics metabolism. J Environ Sci (China) 56:25–35. https://doi.org/10.1016/j.jes.2016.08.022
Liu Q, Tang J, Liu X et al (2019) Vertical response of microbial community and degrading genes to petroleum hydrocarbon contamination in saline alkaline soil. J Environ Sci (China) 81:80–92. https://doi.org/10.1016/j.jes.2019.02.001
Gao Y, Yuan L, Du J et al (2022) Bacterial community profile of the crude oil-contaminated saline soil in the Yellow River Delta Natural Reserve, China. Chemosphere 289:133207. https://doi.org/10.1016/j.chemosphere.2021.133207
Amrutha M, Nampoothiri KM (2022) In silico analysis of nitrilase-3 protein from Corynebacterium glutamicum for bioremediation of nitrile herbicides. J Genet Eng Biotechnol 20:51. https://doi.org/10.1186/s43141-022-00332-5
Ameen F, AlNadhari S, Al-Homaidan AA (2021) Marine microorganisms as an untapped source of bioactive compounds. Saudi J Biol Sci 28:224–231. https://doi.org/10.1016/j.sjbs.2020.09.052
Karthikeyan A, Joseph A, Nair BG (2022) Promising bioactive compounds from the marine environment and their potential effects on various diseases. J Genet Eng Biotechnol 20:14. https://doi.org/10.1186/s43141-021-00290-4
Zhang Y, Liu J, Tang K et al (2015) Genome analysis of Flaviramulus ichthyoenteri Th78(T) in the family Flavobacteriaceae: insights into its quorum quenching property and potential roles in fish intestine. BMC Genomics 16:38. https://doi.org/10.1186/s12864-015-1275-0
Anas A, Nilayangod C, Jasmin C et al (2016) Diversity and bioactive potentials of culturable heterotrophic bacteria from the surficial sediments of the Arabian Sea. 3 Biotech 6:238. https://doi.org/10.1007/s13205-016-0556-x
Soowannayan C, Teja NC, Yatip P et al (2019) Vibrio biofilm inhibitors screened from marine fungi protect shrimp against acute hepatopancreatic necrosis disease (AHPND). Aquaculture 499:1–8. https://doi.org/10.1016/j.aquaculture.2018.09.004
Liu X, Ashforth E, Ren B et al (2010) Bioprospecting microbial natural product libraries from the marine environment for drug discovery. J Antibiot (Tokyo) 63:415–422. https://doi.org/10.1038/ja.2010.56
Xiong ZQ, Wang JF, Hao YY et al (2013) Recent advances in the discovery and development of marine microbial natural products. Mar Drugs 11:700–717. https://doi.org/10.3390/md11030700
Mai Z, Su H, Zhang S (2016) Isolation and characterization of a glycosyl hydrolase family 16 β-agarase from a mangrove soil metagenomic library. Int J Mol Sci 17:e1360. https://doi.org/10.3390/ijms17081360
Schipper C, Hornung C, Bijtenhoorn P et al (2009) Metagenome-derived clones encoding two novel lactonase family proteins involved in biofilm inhibition in Pseudomonas aeruginosa. Appl Environ Microbiol 75:224–233. https://doi.org/10.1128/AEM.01389-08
Li X, Guo J, Dai S et al (2009) Exploring and exploiting microbial diversity through metagenomics for natural product drug discovery. Curr Top Med Chem 9:1525–1535. https://doi.org/10.2174/156802609789909849
Owen J, Robins K, Parachin N et al (2012) A functional screen for recovery of 4’-phosphopantetheinyl transferase and associated natural product biosynthesis genes from metagenome libraries. Environ Microbiol 14:1198–1209. https://doi.org/10.1111/j.1462-2920.2012.02699.x
Wei Y, Zhang L, Zhou Z, Yan X (2018) Diversity of gene clusters for polyketide and nonribosomal peptide biosynthesis revealed by metagenomic analysis of the Yellow Sea sediment. Front Microbiol 9:295. https://doi.org/10.3389/fmicb.2018.00295
Pushpanathan M, Rajendhran J, Jayashree S et al (2012) Identification of a novel antifungal peptide with chitin-binding property from marine metagenome. Protein Pept Lett 19:1289–1296. https://doi.org/10.2174/092986612803521620
Schofield MM, Jain S, Porat D et al (2015) Identification and analysis of the bacterial endosymbiont specialized for production of the chemotherapeutic natural product ET-743. Environ Microbiol 17:3964–3975. https://doi.org/10.1111/1462-2920.1290
Hu Y, Liu Y, Li J et al (2015) Structural and functional analysis of a low-temperature-active alkaline esterase from South China Sea marine sediment microbial metagenomic library. J Ind Microbiol Biotechnol 42:1449–1461. https://doi.org/10.1007/s10295-015-1653-2
De Santi C, Altermark B, Pierechod MM et al (2016) Characterization of a cold-active and salt tolerant esterase identified by functional screening of Arctic metagenomic libraries. BMC Biochem 17:1. https://doi.org/10.1186/s12858-016-0057-x
Lewin A, Zhou J, Pham VTT et al (2017) Novel archaeal thermostable cellulases from an oil reservoir metagenome. AMB Express 7:183. https://doi.org/10.1186/s13568-017-0485-z
Burgess JG (2012) New and emerging analytical techniques for marine biotechnology. Curr Opin Biotechnol 23:29–33. https://doi.org/10.1016/j.copbio.2011.12.007
Trindade M, van Zyl LJ, Navarro-Fernández J et al (2015) Targeted metagenomics as a tool to tap into marine natural product diversity for the discovery and production of drug candidates. Front Microbiol 6:890. https://doi.org/10.3389/fmicb.2015.00890
Lindequist U (2016) Marine-derived pharmaceuticals-challenges and opportunities. Biomol Ther 24:561–571. https://doi.org/10.4062/biomolther.2016.181
Gouda S, Das G, Sen SK et al (2016) Endophytes: a treasure house of bioactive compounds of medicinal importance. Front Microbiol 7:1538. https://doi.org/10.3389/fmicb.2016.01538
Amirzakariya BZ, Shakeri A (2022) Bioactive terpenoids derived from plant endophytic fungi: an updated review (2011–2020). Phytochemistry 197:113130. https://doi.org/10.1016/j.phytochem.2022.113130
Kusari S, Spiteller M (2011) Are we ready for industrial production of bioactive plant secondary metabolites utilizing endophytes? Nat Prod Rep 28:1203. https://doi.org/10.1039/c1np00030f
Challis GL (2008) Genome mining for novel natural product discovery. J Med Chem 51:2618–2628. https://doi.org/10.1021/jm700948z
Scherlach K, Hertweck C (2006) Discovery of aspoquinolones A-D, prenylated quinoline-2-one alkaloids from Aspergillus nidulans, motivated by genome mining. Org Biomol Chem 4:3517–3520. https://doi.org/10.1039/B607011F
Tejesvi MV, Kajula M, Mattila S et al (2011) Bioactivity and genetic diversity of endophytic fungi in Rhododendron tomentosum Harmaja. Fungal Divers 47:97. https://doi.org/10.1007/s13225-010-0087-4
Woźniak M, Grządziel J, Gałązka A et al (2019) Metagenomic analysis of bacterial and fungal community composition associated with Paulownia elongate × Paulownia fortunei. BioRes 14:8511–8529
Parmar S, Li Q, Wu Y et al (2018) Endophytic fungal community of Dysphania ambrosioides from two heavy metal-contaminated sites: evaluated by culture-dependent and culture-independent approaches. Microb Biotechnol 11:1170–1183. https://doi.org/10.1111/1751-7915.13308
Riva V, Mapelli F, Bagnasco A et al (2022) A meta-analysis approach to defining the culturable core of plant endophytic bacterial communities. Appl Environ Microbiol 88:e02537–e02521. https://doi.org/10.1128/aem.02537-21
Joice R, Yasuda K, Shafquat A et al (2014) Determining microbial products and identifying molecular targets in the human microbiome. Cell Metab 20:731–741. https://doi.org/10.1016/j.cmet.2014.10.003
Sharon G, Garg N, Debelius J et al (2014) Specialized metabolites from the microbiome in health and disease. Cell Metab 20:719–730. https://doi.org/10.1016/j.cmet.2014.10.016
Newman DJ, Cragg GM (2012) Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 75:311–335. https://doi.org/10.1021/np200906s
Lemon KP, Armitage GC, Relman DA et al (2012) Microbiota-targeted therapies: an ecological perspective. Sci Transl Med 4:137rv5. https://doi.org/10.1126/scitranslmed.3004183
Lopez CA, Kingsbury DD, Velazquez EM et al (2014) Collateral damage: microbiota-derived metabolites and immune function in the antibiotic era. Cell Host Microbe 16:156–163. https://doi.org/10.1016/j.chom.2014.07.009
Milshteyn A, Schneider JS, Brady SF (2014) Mining the metabiome: identifying novel natural products from microbial communities. Chem Biol 21:1211–1223. https://doi.org/10.1016/j.chembiol.2014.08.006
Koppel N, Balskus EP (2016) Exploring and understanding the biochemical diversity of the human microbiota. Cell Chem Biol 23:18–30. https://doi.org/10.1016/j.chembiol.2015.12.008
Donia MS, Cimermancic P, Schulze CJ et al (2014) A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158:1402–1414. https://doi.org/10.1016/j.cell.2014.08.032
Arnison PG, Bibb MJ, Bierbaum G et al (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. https://doi.org/10.1016/j.cell.2014.08.032
Donia MS, Fischbach MA (2015) Small molecules from the human microbiota. Science 349:1254766. https://doi.org/10.1126/science.1254766
Cohen LJ, Kang HS, Chu J et al (2015) Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc Natl Acad Sci USA 112:E4825–E4834. https://doi.org/10.1073/pnas.1508737112
Tsukimoto M, Nagaoka M, Shishido Y et al (2011) Bacterial production of the tunicate-derived antitumor cyclic depsipeptide didemnin B. J Nat Prod 74:2329–2331. https://doi.org/10.1021/np200543z
Delmont TO, Eren AM, Maccario L et al (2015) Reconstructing rare soil microbial genomes using in situ enrichments and metagenomics. Front Microbiol 6:358. https://doi.org/10.3389/fmicb.2015.00358
McGenity TJ (2018) 2038–When microbes rule the Earth. Environ Microbiol 20:4213–4220. https://doi.org/10.1111/1462-2920.14449
Westmann CA, Alves LF, Silva-Rocha R et al (2018) Mining novel constitutive promoter elements in soil metagenomic libraries in Escherichia coli. Front Microbiol 9:1344. https://doi.org/10.3389/fmicb.2018.01344
Li J, Neubauer P (2014) Escherichia coli as a cell factory for heterologous production of nonribosomal peptides and polyketides. New Biotechnol 31:1–7. https://doi.org/10.1016/j.nbt.2014.03.006
Lorenz P, Eck J (2005) Metagenomics and industrial applications. Nat Rev Microbiol 3:510–516. https://doi.org/10.1038/nrmicro1161
Wilson MR, Zha L, Balskus EP (2017) Natural product discovery from the human microbiome. J Biol Chem 292:8546–8552. https://doi.org/10.1074/jbc.R116.762906
Kalia VC, Gong G, Shanmugam R et al (2022) The emerging biotherapeutic agent. Akkermansia Indian J Microbiol 62:1–10. https://doi.org/10.1007/s12088-021-00993-9
Kalia VC, Shim WY, Patel SKS et al (2022) Recent developments in antimicrobial growth promoters in chicken health: Opportunities and challenges. Sci Total Environ 834:155300. https://doi.org/10.1016/j.scitotenv.2022.155300
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2021H1D3A2A0109705, NRF-2021R1I1A1A01060963, NRF-2020R1I1A1A01073483). This paper was supported by Konkuk University Researcher Fund in 2021. The sponsor(s) had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that there is no actual or potential conflict of interest including any financial, personal or other relationships with other people or organizations within five years of beginning the submitted work that could inappropriately influence, or be perceived to influence, their work.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kalia, V.C., Gong, C., Shanmugam, R. et al. Prospecting Microbial Genomes for Biomolecules and Their Applications. Indian J Microbiol 62, 516–523 (2022). https://doi.org/10.1007/s12088-022-01040-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-022-01040-x