Abstract
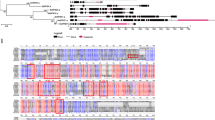
Sugarcane is a major sugar-producing crop, which contributed 80% of the world’s sugar in 2010. Saccarhum officinarum is a domestic species with high sugar content, while, Saccarhum spontaneum is a wild species with stress tolerance. The highly complex polyploid genome of modern sugarcane cultivars arose from the interspecific hybridization between S. officinarum and S. spontaneum. Sucrose synthase (SUS) is a key enzyme for sucrose metabolism in plants, where activity is bidirectional: both synthetic and separate. In this study, nine genomic sequences of S. officinarum and eight genomic sequences of S. spontaneum for five SUS genes were identified. Phylogenetic analysis showed that the Saccharum SUS3 and SUS5 genes were generated from ρ duplication, SUS1 and SUS2 were duplicated after the split of dicot and monocot species, and SUS4 was retained from the last common ancestor before the origination of Angiospermae. The gene structure and Ka/Ks analysis suggested the functional constraint of SUS genes in the two Saccharum species. Gene expression based on RNA-seq analysis revealed that SUS1was dominantly expressed in source tissues including the internodes and the basal zone of the leaves, SUS2 was detectable in all tissues examined, and the remaining three SUS genes were expressed at low levels in the examined tissues, indicating SUS1 is the key member involved in sucrose accumulation. In addition, SUS genes were observed to be present at higher expression levels in S. officinarum than in S. spontaneum, while SUS2 presented different expression patterns during the circadian rhythm in S. spontaneum and S. officinarum, suggesting the two SUS genes contribute to the differential sugar levels in these species. Our comprehensive study in Saccharum provides the foundations for further functional studies of the SUS gene family.






Similar content being viewed by others
References
An X, Chen Z, Wang J, Ye M, Ji L, Wang J, Liao W, Ma H (2014) Identification and characterization of the Populus sucrose synthase gene family. Gene 539:58–67. https://doi.org/10.1016/j.gene.2014.01.062
Baier MC, Keck M, Godde V, Niehaus K, Kuster H, Hohnjec N (2010) Knockdown of the symbiotic sucrose synthase MtSucS1 affects arbuscule maturation and maintenance in mycorrhizal roots of Medicago truncatula. Plant Physiol 152:1000–1014. https://doi.org/10.1104/pp.109.149898
Barratt DHP, Barber L, Kruger NJ, Smith AM, Wang TL, Martin C (2001) Multiple, distinct isoforms of sucrose synthase in pea. Plant Physiol 127:655–664. https://doi.org/10.1104/pp.010297
Baud S, Vaultier MN, Rochat C (2004) Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J Exp Bot 55:397–409. https://doi.org/10.1093/jxb/erh047
Bieniawska Z, Barratt D, Garlick A, Thole V, Nj MC, Zrenner R, Smith A (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J Cell Mol Biol 49:810–828
Bläsing OE, Stitt M (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17:3257–3281
Buxton DR, Redfearn DD (1997) Plant limitations to fiber digestion and utilization. J Nutr 127:814S–818S
Chalhoub B et al (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345:950–953
Chen A, He S, Li F, Zhao L, Ding M, Liu Q, Rong J (2012) Analyses of the sucrose synthase gene family in cotton: structure, phylogeny and expression patterns. BMC Plant Biol 12:85
Chen Z-L, Gui Y-Y, Qin C-X, Wang M, Liao Q, Huang D-L, Li Y-R (2015) Isolation and expression analysis of sucrose synthase gene (ScSuSy4) from sugarcane. Sugar Tech 18:134–140. https://doi.org/10.1007/s12355-015-0372-3
Chourey PS, Taliercio EW, Carlson SJ, Ruan YL (1998) Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol Gen Genet MGG 259:88–96
Duncan KA, Hardin SC, Huber SC (2006) The three maize sucrose synthase isoforms differ in distribution, localization, and phosphorylation. Plant Cell Physiol 47:959–971. https://doi.org/10.1093/pcp/pcj068
Flagel LE, Wendel JF (2009) Gene duplication and evolutionary novelty in plants. New Phytol 183:557–564
Fu H, Park WD (1995) Sink- and vascular-associated sucrose synthase functions are encoded by different gene classes in potato. Plant Cell 7:1369–1385
Fujii S, Hayashi T, Mizuno K (2010) Sucrose synthase is an integral component of the cellulose synthesis machinery. Plant Cell Physiol 51:294–301. https://doi.org/10.1093/pcp/pcp190
Geigenberger P, Stitt M (1993) Sucrose synthase catalyses a readily reversible reaction in vivo in developing potato tubers and other plant tissues. Planta 189:329–339
Harada T, Satoh S, Yoshioka T, Ishizawa K (2005) Expression of sucrose synthase genes involved in enhanced elongation of pondweed (Potamogeton distinctus) turions under anoxia. Ann Bot 96:683–692
Hirose T, Scofield GN, Terao T (2008) An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci 174:534–543. https://doi.org/10.1016/j.plantsci.2008.02.009
Hohnjec N, Perlick AM, Pühler A, Küster H (2003) The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol Plant-Microbe Interact 16:903–915
Irvine JE (1999) Saccharum species as horticultural classes. Theor Appl Genet 98:186–194
Jiao Y, Paterson AH (2014) Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 26:2792–2802
Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, dePamphilis CW (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100
Kleczkowski L, Kunz S, Wilczynska M (2010) Mechanisms of UDP-glucose synthesis in plants. Crit Rev Plant Sci 29:191–203. https://doi.org/10.1080/07352689.2010.483578
Klotz KL, Finger FL, Shelver WL (2003) Characterization of two sucrose synthase isoforms in sugarbeet root. Plant Physiol Biochem 41:107–115. https://doi.org/10.1016/s0981-9428(02)00024-4
Lam E et al (2009) Improving sugarcane for biofuel: engineering for an even better feedstock. GCB Bioenergy 1:251–255. https://doi.org/10.1111/j.1757-1707.2009.01016.x
Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, Reidel EJ, Turgeon R, Liu P, Sun Q, Nelson T, Brutnell TP (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42:1060–1067. https://doi.org/10.1038/ng.703
Lingle SE, Dyer JM (2001) Cloning and expression of sucrose synthase-1 cDNA from sugarcane. J Plant Physiol 158:129–131
Lutfiyya LL, Xu N, D’Ordine RL, Morrell JA, Miller PW, Duff SMG (2007) Phylogenetic and expression analysis of sucrose phosphate synthase isozymes in plants. J Plant Physiol 164:923–933
Ming R et al (2015) The pineapple genome and the evolution of CAM photosynthesis. Nat Genet 47:1435–1442
Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci U S A 101:9903–9908
Schaffer AA, Sagee O, Goldschmidt EE, Goren R (2010) Invertase and sucrose synthase activity, carbohydrate status and endogenous IAA levels during Citrus leaf development. Physiol Plant 69:151–155
Schmalstig JG, Hitz WD (1987) Contributions of sucrose synthase and Invertase to the metabolism of sucrose in developing leaves : estimation by alternate substrate utilization. Plant Physiol 85:407–412. https://doi.org/10.1104/pp.85.2.407
Sun J, Loboda T, Sung SJS, Black CC (1992) Sucrose synthase in wild tomato, Lycopersicon chmielewskii, and tomato fruit sink strength. Plant Physiol 98:1163–1169. https://doi.org/10.1104/pp.98.3.1163
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. https://doi.org/10.1093/molbev/msr121
Tang GQ, Sturm A (1999) Antisense repression of sucrose synthase in carrot (Daucus carota L.) affects growth rather than sucrose partitioning. Plant Mol Biol 41:465–479
Thompson JD, Gibson TJ, Higgins DGJCPB (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2:Unit 2.3
Wang X, Shi X, Hao B, Ge S, Luo J (2005) Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol 165:937–946
Wang J, Roe B, Macmil S, Yu Q, Murray JE, Tang H, Chen C, Najar F, Wiley G, Bowers J, van Sluys MA, Rokhsar DS, Hudson ME, Moose SP, Paterson AH, Ming R (2010) Microcollinearity between autopolyploid sugarcane and diploid sorghum genomes. BMC Genomics 11:261
Wang Z, Wei P, Wu M, Xu Y, Li F, Luo Z, Zhang J, Chen A, Xie X, Cao P, Lin F, Yang J (2015) Analysis of the sucrose synthase gene family in tobacco: structure, phylogeny, and expression patterns. Planta 242:153–166. https://doi.org/10.1007/s00425-015-2297-1
Wang Y, Hua X, Xu J, Chen Z, Zhang J (2019) Comparative genomics revealed the gene evolution and functional divergence of magnesium transporter families in saccharum. BMC Genomics 20:83 https://doi.org/10.1186/s12864-019-5437-3
Xiao X, Tang C, Fang Y, Yang M, Zhou B, Qi J, Zhang Y (2014) Structure and expression profile of the sucrose synthase gene family in the rubber tree: indicative of roles in stress response and sucrose utilization in the laticifers. FEBS J 281:291–305
Xu G, Guo C, Shan H, Kong H (2012) Divergence of duplicate genes in exon-intron structure. Proc Natl Acad Sci 109:1187–1192
Zhang Z, Li J, Zhao XQ, Wang J, Wong KS, Yu J (2006) KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4:259–263
Zhang D, Xu B, Yang X, Zhang Z, Li B (2011) The sucrose synthase gene family in Populus : structure, expression, and evolution. Tree Genet Genomes 7:443–456
Zhang J, Nagai C, Yu Q, Pan YB, Ayala-Silva T, Schnell RJ, Comstock JC, Arumuganathan AK, Ming R (2012) Genome size variation in three Saccharum species. Euphytica 185:511–519. https://doi.org/10.1007/s10681-012-0664-6
Zhang J, Arro J, Chen Y, Ming R (2013) Haplotype analysis of sucrose synthase gene family in three Saccharum species. BMC Genomics 14:314. https://doi.org/10.1186/1471-2164-14-314
Zhang J et al (2018) Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat Genet 50:1565–1573. https://doi.org/10.1038/s41588-018-0237-2
Zhenglong G, Steinmetz LM, Xun G, Curt S, Davis RW, Wen-Hsiung L (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 421:63–66
Zou C, Lu C, Shang H, Jing X, Cheng H, Zhang Y, Song G (2013) Genome-wide analysis of the Sus gene family in cotton. J Integr Plant Biol 55:643–653
Zrenner R, Salanoubat M, Willmitzer L, Sonnewald U (1995) Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). Plant J 7:97–107
Acknowledgements
This project was supported by grants from National key research and development program (2018YFD1000104), the 863 program (2013AA100604), NSFC (31201260, 31760413 and 31660420), Science and Technology Major Project of Guangxi (AA17202025) and Fujian Provincial Department of Education (No. JA12082). We are grateful for language editing the by Irene Lavagi.
Author information
Authors and Affiliations
Contributions
JZ designed the experiments. JZ and YS conceived the study. YS, HX, QS, JL,YW, XH, WY, QYand, RM performed the experiments and analyzed the data. JZ and YS wrote the manuscript. All authors read and approved the final article.
Corresponding author
Additional information
Communicated by: Hongwei Cai
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplemental Table 1
(DOCX 23 kb)
Supplemental Table 2
(XLSX 87 kb)
Supplemental Table 3
(XLSX 87.7 kb)
Supplemental Figure 1
(PNG 62464 kb)
Rights and permissions
About this article
Cite this article
Shi, Y., Xu, H., Shen, Q. et al. Comparative Analysis of SUS Gene Family between Saccharum officinarum and Saccharum spontaneum. Tropical Plant Biol. 12, 174–185 (2019). https://doi.org/10.1007/s12042-019-09230-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-019-09230-6