Abstract
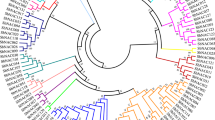
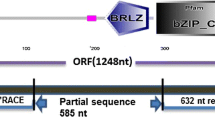
In this study, we show that NAC-like transcription factor (TF) has 90% sequence similarity with cDNA of the OsNac5 gene present in the NCBI database. Phylogenetic analysis of the NAC gene family was performed with inclusion of the highly diverse C-terminal sequences. We report that this gene is also found to be orthologous to Glycine max NAC8, NAC2, Triticum aestivum NAC6 and paralogous to OsNAC6. mRNA was purified from five recombinant inbred lines (RILs) and parents of rice at three different stages of grain filling under aerobic conditions, with grain protein content (GPC) spanning from 4 to 14%. The NAC-like TF encoding a protein was found to be upregulated at the \(\hbox {S}_{2}\) stage in the leaf (3.9-fold) and panicle (1.84-fold) of parent HPR14 and in five RILs (1.9 to 4.51-fold in leaves and 0.47 to 3.2-fold in panicles). Expression analysis of the NAC-like TF encoding a protein for the rice gene was found to be upregulated at the \(\hbox {S}_{2}\) stage in the leaf and panicle of parental line HPR14 and RILs with high protein content.





Similar content being viewed by others
References
Aida M., Ishida T., Fukaki H., Fujisawa H. and Tasaka M. 1997 Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857.
Barneix A. J. and Guitman M. R. 1993 Leaf regulation of the nitrogen concentration in the grain of wheat plants. J. Exp. Bot. 44, 1607–1612.
Christianson J. A., Wilson I. W., Llewellyn D. J. and Dennis E. S. 2009 The low oxygen induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis thaliana seeds following low-oxygen treatment. Plant Physiol. 149, 1724–1738.
Edgar R. C. 2004 MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Ernst H. A., Olsen A. N., Skriver K., Larsen S. and Leggio L. L. 2004 Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5, 297–303.
Fang Y., You J., Xie K., Xie W. and Xiong L. 2008 Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genom. 280, 547–563.
Felsenstein J. 1989 PHYLIP – phylogeny inference package (Version 3.2). Cladistics 5, 163–166.
Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu, K., Ohme-Takagi M. et al. 2004 A dehydration induced NAC protein, RD26, is involved in a novel ABA-dependent stress signaling pathway. Plant J. 39, 863–876.
Greve K., La Cour T., Jensen M. K., Poulsen F. M. and Skriver K. 2003 Interactions between plant RING-H2 and plant-specific NAC (NAM/ATAF1/2/CUC2) proteins: RING-H2 molecular specificity and cellular localization. Biochem. J. 371, 97–108.
Hegedus D., Yu M., Baldwin D., Gruber M., Sharpe A., Parkin I. et al. 2003 Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol. Biol. 53, 383–397.
Hollander M. and Wolfe D. A. 1973 Nonparametric statistical methods, pp. 503. Wiley, New York.
Hu H., Dai M., Yao J., Xiao B., Li X., Zhang Q. et al. 2006 Overexpressing a NAM, ATAF and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. 103, 12987–12992.
Hu H., You J., Fang Y., Zhu X., Qi Z. and Xiong L. 2008 Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 67, 169–181.
Jain M., Nijhawan A., Arora R., Agarwal P., Ray S., Sharma P. et al. 2007 F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143, 1467–1483.
Jeong J. S., Park Y. T., Jung H., Park S. H. and Kim J. K. 2009 Rice NAC proteins act as homodimers and heterodimers. Plant Biotechnol. Rep. 3, 127–134.
Jeong J. S., Kim Y. S., Baek K. H., Jung H., Ha S. H., Choi Y. D. et al. 2010 Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–197.
Jeong J. S., Kim Y. S., Redillas M. C., Jang G., Jung H., Bang S. W. et al. 2013 OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11, 101–114.
Juliano B. O. 1971 A simplified assay for milled-rice amylose. Cereal Sci. Today 16, 334, 340, 360.
Kade M. A., Barneix J., Olmos S. and Dubcovsky J. 2005 Nitrogen uptake and remobilization in tetraploid Langdon durum wheat and a recombinant substitution line with the high grain protein gene GPC-B1. Plant Breed. 124, 343–349.
Kawakatsu T., Wang S., Wakasa Y. and Takaiwa F. 2010 Increased lysine content in rice grains by over-accumulation of BiP in the endosperm. Biosci. Biotechnol. Biochem. 74, 2529–2531.
Krishnan A., Guiderdoni E., An, G., Hsing Y. I., Han C. D. and Lee M. C. 2009 Mutant resources in rice for functional genomics of the grasses. Plant Physiol. 149, 165–170.
Li W. and Godzik A. 2006 Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659.
Lim P. O., Kim H. J. and Nam H. G. 2007 Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136.
Martre P., Porter J. R., Jamieson P. D. and Triboi E. 2003 Modeling grain nitrogen accumulation and protein composition to understand the sink/source regulations of nitrogen remobilization for wheat. Plant Physiol. 133, 1959–1967.
McGinnis S. and Madden T. L. 2004 BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32, 20–25.
Nakashima K., Tran L. S., Van Nguyen D., Fujita M., Maruyama K., Todaka, D. et al. 2007 Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630.
Ooka H., Satoh K., Doi K., Nagata T., Otomo Y., Murakami K. et al. 2003 Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 20, 239–247.
Peng H., Cheng H. Y., Chen C., Yu X. W., Yang J. N., Gao W. R. et al. 2009 A NAC transcription factor gene of chickpea (Cicer arietinum), CarNAC3, is involved in drought stress response and various developmental processes. J. Plant Physiol. 166, 1934–1945.
Pinheiro G. L., Marques C. S., Costa M. D., Reis P. A., Alves M. S., Carvalho C. M. et al. 2009 Complete inventory of soybean NAC transcription factors: sequence conservation and expression analysis uncover their distinct roles in stress response. Gene 444, 10–23.
Sablowski R. W. and Meyerowitz E. M. 1998 A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92, 93–103.
Shen H., Yin Y. B., Chen F., Xu Y. and Dixon R. A. 2009 A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. Bioenergy Res. 2, 217–232.
Song S. Y., Chen Y., Chen J., Dai X. Y. and Zhang W. H. 2011 Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234, 331–345.
Souer E, Van Houwelingen A, Kloos D, Mol J. and Koes R. 1996 The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170.
Sperotto R. A., Ricachenevsky F. K., Duarte G. L., Boff T., Lopes K. L. and Sperb E. R. 2009 Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 230, 985–1002.
Sperotto R. A., Boff T., Duarte G. L., Santos L. S., Grusak M. A. and Fett J. P. 2010 Identification of putative target genes to manipulate Fe and Zn concentrations in rice grains. J. Plant Physiol. 167, 1500–1506.
Takasaki H., Maruyama K., Kidokoro S., Ito Y., Fujita Y., Shinozaki K. et al. 2010 The abiotic stress responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genom. 284, 173–183.
Tian Z., Qian Q., Liu Q., Yan M., Liu X. and Yan C. 2009 Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. USA 106, 21760–21765.
Tran L. S., Nakashima K., Sakuma Y., Simpson S. D., Fujita Y., Maruyama K. et al. 2004 Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498.
Uauy C., Distelfeld A., Fahima T., Blechl A. and Dubcovsky, J. 2006 A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314, 1298–1301.
Verma P. S. and Agarwal V. K. 2010 Genetics, 3rd edition. S. Chand publication.
Viraktamath B. C., Bentur J. S., Rao K. V. and Mangal S. 2011 Vision 2030, Directorate of Rice Research, Hyderabad.
Xie Q., Frugis G., Colgan D. and Chua N. H. 2000 Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036.
Yuan S. J. and Neal Stewart C. N. 2005 Real-time PCR statistics. PCR Encycl. 1, 101127–101149.
Zhang W., Bi J., Chen L., Zheng L., Ji S., Xia Y. et al. 2008 QTL mapping for crude protein and protein fraction contents in rice (Oryza sativa L.). J. Cereal Sci. 48, 539–547.
Zheng X., Chen B., Lu G. and Han B. 2009 Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem. Biophys. Res. Commun. 379, 985–989.
Acknowledgements
We thank the Department of Biotechnology (DBT), New Delhi, Government of India, for funding this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Arun Joshi
Rights and permissions
About this article
Cite this article
Sharma, G., Upadyay, A.K., Biradar, H. et al. OsNAC-like transcription factor involved in regulating seed-storage protein content at different stages of grain filling in rice under aerobic conditions. J Genet 98, 18 (2019). https://doi.org/10.1007/s12041-019-1066-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-019-1066-5