Abstract
Loss of E3 ubiquitin ligase RING finger protein 8 (RNF8) may lead to neuronal DNA damage and apoptosis. In order to expand on our knowledge on the mechanistic basis underlying neuronal death in ischemic stroke, the present study sought to investigate the potential role of E3 ubiquitin ligase RNF8 on ischemic stroke and explore the underlying downstream mechanism. Middle cerebral artery occlusion (MCAO) in mice and oxygen–glucose deprivation/reoxygenation (OGD/R) in neurons were induced to simulate an ischemic stroke environment. It was found that downregulation of RNF8 and Reelin occurred in MCAO mice and OGD/R-exposed neurons. Silencing of RNF8 enhanced the MCAO-induced neuronal apoptosis and oxidative stress. Mechanistically, RNF8 enhanced the ubiquitination and degradation of HDAC2, thus attenuating OGD/R-induced neuronal apoptosis and oxidative stress. Moreover, HDAC2 inhibited Reelin expression through deacetylation of H3K27me3 in its promoter, causing reduced glycogen synthase kinase-3beta (GSK3β)-Ser9 phosphorylation and the resultant elevated GSK3β activity. By this mechanism, RNF8 alleviated ischemic stroke. Coherently, this study suggests that RNF8 plays a neuroprotective effect against ischemic stroke by downregulating HDAC2 expression and inducing Reelin-induced GSK3β inhibition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stroke is the leading cause of mortality and disability worldwide, with ischemic stroke being the most prevalent form of stroke [1]. Patients affected by the coronavirus disease 2019 (COVID-19) may develop coagulopathy with thromboembolic complications, including ischemic stroke [2]. SARS-CoV-2 infection can increase the risk of ischemic stroke since it may induce an inflammatory response and trigger a hypercoagulation state [3]. Nowadays, treatment approaches comprise of medical, endovascular, and surgical strategies [4], but the vast majority of survivors succumb to the risk of recurrence and adverse symptoms like neurocognitive [5] and somatosensory deficits [6, 7]. The understanding of the molecular mechanisms underlying neurophysiological processes following the stroke is pivotal for the development of new therapies and treatments to improve neuronal recovery [8].
RING finger protein 8 (RNF8) is known as an E3 ubiquitin ligase, with recognized participation in the alleviation of ischemic stroke [9,10,11]. Meanwhile, the loss of RNF8 contributes to neuronal pathology and cognitive decline, accelerating the resultant degeneration of neurons [12]. Prediction results from the UbiBrowser indicated that histone deacetylase 2 (HDAC2) can be used as the substrate of RNF8. Histone deacetylases (HDACs) are widely known as a family of enzymes, that possess the ability to catalyze the removal of acetyl groups from the acetyl-lysine residues in histone and non-histone proteins [13], and often function as epigenetic regulators involved in the pathological processes of malignancies, neurological diseases, and immune disorders [14]. Recent data has further identified HDAC2 to serve as a potential target for anti-stroke therapy, owing to the fact that HDAC2 inhibition restricted neuronal apoptosis in the penumbra tissue after ischemic stroke [15]. Furthermore, treatment with HDAC inhibitors (valproic acid [VPA] and suberoylanilide hydroxamic acid) can restore the expression of Reelin in pancreatic cancer cells [16].
Reelin is an extracellular matrix protein secreted in embryonic and adult brain, and known to be essential for critical processes like neuron migration and neuronal plasticity [17]. Interestingly, Reelin upregulation can also confer protection against stroke-induced brain injury, attributed to its ability to increase cell survival following oxidative injury and reduce infarct volume and neurological deficit [18].
Of note, a previous study has also demonstrated that the augmentation of glycogen synthase kinase-3beta (GSK3β) activity upon Reelin knockout in mouse models of Alzheimer’s disease [19]. This is particularly important as the inhibition of GSK3β is known to delay ischemic stroke by reducing autophagy and increasing the expression of phosphorylated (p)-mammalian target of rapamycin, p-GSK3β, and β-catenin in oxygen–glucose deprivation/reoxygenation (OGD/R) neurons [20]. Of note, Wang et al. recently reported that suppression of GSK3β activity can aggravate cerebral ischemia/reperfusion-induced injury by enhancing cell autophagy [21].
In light of the previous evidence, the current study aims to explore the potential neuroprotective role of RNF8 in ischemic stroke and investigate the underlying mechanism involving the HDAC2/Reelin/GSK3β signaling on the basis of middle cerebral artery occlusion (MCAO) mouse models and OGD/R-induced cell models.
Materials and Methods
Ethics Statement
The current study was carried out under the approval of the Ethics committee of the Affiliated Hospital of Guizhou Medical University (approval number: 2020-046A), and conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
Microarray-Based Gene Expression Profiling
Ischemic stroke-related datasets GSE23160, GSE58720, and GSE102541 were downloaded from the Gene Expression Omnibus (GEO) database. GSE23160 contains 8 brain tissue samples of sham-operated mice and 24 brain tissue samples of MCAO mouse models. GSE58720 contains 3 brain tissue samples of sham-operated mice and 3 brain tissue samples of MCAO mouse models. GSE102541 contains 3 blood samples from healthy controls and 6 blood samples from ischemic stroke patients. R “limma” package was applied for differential analysis to identify the differentially expressed genes with p value < 0.01 as the threshold. Then, 514 E3 ubiquitin ligases (species: Homo sapiens, Mus musculus) were obtained from the iUUCD database and intersected with the differentially expressed genes.
Establishment of MCAO-Induced Mouse Ischemic Stroke Models
RNF8floxed/floxed C57BL/6 and Nestin-CRE+/− C57BL/6 mice (purchased from Jackson Laboratory) were housed in a specific pathogen-free environment. RNF8 conditional knockout (RNF8−/−) mice were constructed as follows: male RNF8floxed/+ CRE+ and female RNF8floxed/+ CRE− mice were obtained by hybridizing male Nestin-CRE+/− and female RNF8floxed/floxed mice. These two mice were hybridized to generate the experimental cohort: male RNF8floxed/Δcre+ mice were RNF8−/− mice in the nervous system, and male wild-type (WT) mice (+ / + cre + , + / + cre − , or floxed/ + cre −). Genotyping was determined by the DNA isolated from mouse tail-cutting and PCR analysis for WT, floxed, and deletion alleles [22]. RNF8 flox alleles were genotyped using the following PCR primers: RNF8 forward sequence of 5′-GGTTACCACTCCATAACCATCTGTACG-3′ and reverse sequence of 5′-CAGAAGGTAGCAACAGAACACGACG-3′ [23].
Six-to-eight-week-old male WT mice (weighing 18–22 g) or RNF8−/− mice (weighing 18–22 g) were housed at 22 °C with 70% humidity, under a 12-h light/dark cycle and given ad libitum access to food and water. Prior to MCAO, the mice were anesthetized using an isoflurane respiratory anesthesia machine (80% N2O, 20% O2 containing 2% isoflurane for induction of anesthesia, 1.5% isoflurane used for maintaining anesthesia). The intraluminal filament method was applied for MCAO establishment. The common carotid artery, internal carotid artery, and external carotid artery were separated, and a silicone-coated 6–0 nylon suture was inserted from the external carotid artery, through the internal carotid artery, and then gently inserted into the middle cerebral artery. The success of occlusion was determined by monitoring the decrease in surface cerebral blood flow (CBF) to 10% of baseline CBF using a laser Doppler flowmeter (Moor Instruments, Devon, UK). Following MCAO induction for 1 h, the suture was withdrawn and reperfusion was followed. A recovery of CBF to more than 70% of basal CBF within 10 min was indicative of successful reperfusion. The sham-operated mice underwent the same procedures except for suture insertion into the artery. Other physiological parameters were monitored, and the body temperature was maintained at 37.0 ± 0.5 °C using an electric heating pad.
Later, WT mice (n = 24) serving as normal control and RNF8−/− mice following MCAO (n = 24) were both infected with adeno-associated viruses expressing vector and Reelin, with 8 mice used for each treatment. At 72 h after MCAO induction, evaluation was conducted by two observers independently using neurological function scoring. Mice with scores 0 and 8 were excluded, and n = 8 was guaranteed.
Adeno-Associated Virus Vector Construction and Transduction
Single-stranded adeno-associated viruses (ssAAV2-Reelin, 1.5 × 109 vg/mL) were purchased from Shanghai Genechem Co., Ltd. (Shanghai, China). The sequence was designed according to mouse Reelin and finalized by Genechem. Briefly, clone was performed based on the mRNA sequence Reelin (XM_031455859) with the mouse genome as the template. The correct clonal products were sequenced, the results of which turned out to be the same with Reelin promoter region registered on GenBank. Next, the virus solution was dissolved in diethyl pyrocarbonate solution, 2 μL of which was delivered to the infarct site (anterior–posterior, 0 mm from the anterior; medial–lateral, 1.5 mm; dorsal-abdominal, 1.3 mm) by stereotactic injection at a speed of 2 nL/s. The needle was kept for 10 min to ensure even virus distribution after injection.
Neurological Deficit Scores and Cerebral Infarct Volume
After 72 h of MCAO, neurological deficit scores were assessed by two observers independently. The scoring system was as follows: 0 point indicated absence of neurological deficit; 1 point indicated flexion of left forelimb with tail suspension or incomplete extension of right forepaw; 2 points indicated left shoulder adduction during tail suspension; 3 points indicated a reduction of resistance to left push; 4 points indicated spontaneous movement in all directions, only turning left when pulled by tail; 5 points indicated hovering only to the left or spontaneous walking; 6 points indicated walking only when stimulated; 7 points indicated no response to stimulation; and 8 points indicated stroke-related death.
After neurological deficit evaluation, the mice were euthanized by carbon dioxide inhalation for evaluation of mouse cerebral infarct volume. After 10 min of precooling in a fridge at − 20 °C, the brain was taken out and sliced into 1-mm-thick coronal sections, which were immersed in 2% TTC staining solution (17,779-10X10ML-F, Sigma-Aldrich, St Louis, MO) at 37 °C for 15 min. Red coloration was indicative to normal brain tissues, while light gray coloration indicated infarct tissues. The Image-Pro Plus 6.0 software (Media Cybernetics, Bethesda, MD) was employed for section photograph and analysis. Infarct volume = the sum of infarct area × thickness of brain tissue sections.
Hematoxylin–Eosin (HE) and Nissl Staining Assays
HE staining kits (C0105S, Beyotime, Shanghai, China) and Nissl staining solution (G1430, Solarbio, Beijing, China) were used in this study. The brain tissues were fixed with 4% paraformaldehyde and sliced into sections. Subsequently, the sections were stained with hematoxylin for 5–10 min or Nissl dyeing solution for 1 h at 56 °C. For HE staining, the sections were subsequently stained with eosin for 30 s–2 min, while for Nissl staining, the sections were immersed in Nissl differentiation solution until the background was almost colorless under a microscope. The sections were observed under an inverted microscope (IX73, Olympus, Japan). Three sections were taken from each mouse brain tissue section, with three images obtained, and the positive cells were counted with ImageJ software.
Primary Cortical Neuron Culture and Treatment
Primary neurons were extracted from cerebral cortices of 1-day-old male C57BL/6 J mice. Brain tissues were sliced into sections, detached using 0.25% trypsin (25,200–072, Gibco, Carlsbad, CA), allowed to stand at 37 °C for 20 min, and gently ground. Cells were spread on 6-well plates coated with polylysine (100 μg/mL, P4707, Sigma-Aldrich) and cultured with nerve basic medium (21,103–049, Gibco) in a humidified 5% CO2 incubator at 37 °C. After 8 h of incubation, the medium was changed and then renewed every other day and a half. For OGD/R exposure, after 7–9 days of culture, neurons were rinsed with phosphate buffer saline (PBS) thrice, added with glucose-free Dulbecco’s modified Eagle’s medium (11,966–025, Gibco), and placed in an anaerobic chamber (5% CO2 and 95% N2) at 37 °C for 2 h of culture. Next, the medium was replaced by normal medium and further cultured for 12 h in a humidified 5% CO2 incubator at 37 °C for reoxygenation. The control cells were cultured in complete medium under normal conditions.
Short hairpin RNA (shRNA) (shNC), shRNA-1 against RNF8 (shRNF8-1), shRNF8-2, shHDAC2-1, shHDAC2-2, vector (control for over-expressed lentivirus), RNF8 (NM_003958), HDAC2, and Reelin plasmids were constructed, with shRNA sequences listed in Table S1. Briefly, the mRNA sequences of RNF8 (NM_003958), HDAC2 (NM_001527), and Reelin (XM_031455859) were first determined, and plasmid design was performed by Genechem. pLKD-Ubc-eGFP-U6 was used as the interference vector, and pLenti-Ubc-EGFP was selected as the over-expression vector, both of which were purchased from Genechem Co., Ltd. (Shanghai, China). According to the mouse gene sequence, the transduction sequences were cloned into the corresponding vectors to obtain corresponding lentivirus. Next, the cells were seeded at a density of 1 × 105 cells/well [26], and when cell confluence reached approximately 50–70%, cells were transduced with the virus harboring shNC, shRNF8-1, shRNF8-2, shHDAC2-1, shHDAC2-2, vector, RNF8, HDAC2, and Reelin. The culture solution in the plate was removed and then renewed with solution harboring virus. Following 2-h incubation in an incubator, the cell culture solution was renewed with normal cell culture solution for further incubation in an incubator for another 48 h.
Protein synthesis inhibitor cycloheximide (CHX) (SC0353) and proteasome inhibitor MG132 (SC0353) were both purchased from Beyotime. The mouse primary cortical neurons were treated with shNC or shRNF8 for 48 h, and then with CHX (50 μg/mL). Cells were collected at 0, 3, 6, and 9 h after administration and HDAC2 protein expression was detected. The mouse primary cortical neurons were treated with vector or RNF8 for 36 h, and then with 10 μM MG132 for 8 h. The cells were then collected and the protein expression of RNF8 and HDAC2 was detected [24,25,26].
Immunofluorescence Staining
The brain tissues of mice were fixed with 4% paraformaldehyde, and then sliced into 40-μm-thick sections. Following antigen retrieval with sodium citrate buffer, the sections were blocked with 10% normal donkey serum and immunostained with primary rabbit antibody to HDAC2 (ab32117, 1:250, Abcam Inc., Cambridge, UK) overnight at 4 °C. The following day, the sections were incubated with secondary antibody goat anti-rabbit immunoglobulin G (IgG) (ab150078, 1:100, Abcam) coupled with Alexa Fluor 555 at room temperature for 1 h. The cells were stained using DAPI (C1006, Beyotime) and observed under a fluorescence microscope (BX63, Olympus). Three sections were taken from each mouse brain tissue section, with three images obtained, and the positive cells counted with ImageJ software.
Co-immunoprecipitation (Co-IP) Assay
The cells were incubated with 0.5 mL of precooled IP lysis buffer (P0013, Beyotime) on ice for 5 min. Next, the cells were ultrasonicated and then centrifuged at 14,000 × g and 4 °C for 10 min. Thereafter, the supernatant (cell lysate) was incubated overnight at 4 °C with anti-RNF8 antibody (14,112–1-AP, 1:50, ProteinTech, Chicago, IL) or anti-HDAC2 antibody (ab32117, 1:50, Abcam), with non-specific IgG antibody (ab172730, 1:50, Abcam) serving as NC. The cell lysate was then supplemented with 5 μL protein A/G magnetic beads (78,610, Pierce, Rockford, IL). The obtained precipitate was then rinsed with 0.5 mL Tris-buffered saline (TBS) solution and centrifuged at 12,000 × g for 1 min. The sample was finally analyzed by western blot analysis.
TUNEL Assay
According to the instructions of red fluorescent TUNEL Kit (C1089, Beyotime), tissues were fixed with 4% paraformaldehyde and incubated with 0.5% Triton X-100 PBS at room temperature for 5 min. Next, 50 μL TUNEL detection solution was added to the tissue sections and incubated at 37 °C for 60 min in dark conditions. Next, the tissues were sealed with anti-fade mounting medium (P0126, Beyotime) and observed under a FV-1000/ES confocal microscope in five randomly selected visual fields.
For cell treatment, cells were fixed with 4% paraformaldehyde for 30 min and incubated with PBS containing 0.3% Triton X-100 for 5 min at room temperature. Then, 50 μL TUNEL detection solution was added to the cells and incubated at 37 °C for 60 min under dark conditions. Next, the cells were sealed with anti-fade mounting medium and observed under a FV-1000/ES confocal microscope in five randomly selected visual fields. For animal experiments, three sections were taken from each mouse brain tissue section, with three images obtained. For in vitro cell culture experiments, each experiment was repeated three times to obtain three images and the positive cells were counted with ImageJ software.
RNA Isolation and Quantitation
Total RNA content was extracted from tissues using the TRIzol reagent (16,096,020, Invitrogen, Carlsbad, CA), which was then reverse-transcribed into complementary DNA (cDNA) using a reverse transcription kit (RR047A, Takara, Japan). Subsequently, reverse transcription quantitative PCR (RT-qPCR) was conducted in accordance to the instructions of TaqMan Gene Expression Assays protocol (Applied Biosystems, Foster City, CA) using an ABI 7500 instrument (Applied Biosystems), with three replicates set. All primer sequences were designed using the NCBI and are listed in Table S2. The fold changes were calculated using the method of 2−ΔΔCt with GAPDH serving as the internal control.
Western Blot Analysis
Total protein content was extracted from cells lysing with radio-immunoprecipitation assay lysis buffer (P0013C, Beyotime) containing protein inhibitor. Next, the protein concentration was determined using a bicinchoninic acid kit (20201ES76, YEASEN Biotechnology Co., Ltd., Shanghai, China). After separation by sodium dodecyl sulfate polyacrylamide gel electrophoresis, the proteins were transferred onto a polyvinylidene fluoride membrane (IPVH85R, Millipore, Germany). Subsequently, the membrane was blocked with 5% bovine serum albumin at room temperature for 1 h and then underwent overnight incubation at 4 °C with primary antibodies against RNF8 (ab128872, 1:1000), HDAC2 (ab32117, 1:2000), Reelin (ab78540, 1:1000), GSK3β (ab32391, 1:1000), p-GSK3β-Ser9 (ab131097, 1:2000), and β-actin (ab8226, 1:3000). The following day, the membrane was re-probed with horseradish peroxidase-labeled secondary goat anti-rabbit IgG (ab6721, 1:10,000) or rabbit anti-mouse IgG (ab6728, 1:10,000) at room temperature for 1 h. The aforementioned antibodies were purchased from Abcam. Afterwards, the membrane was visualized using luminescent liquid and the protein bands were quantified with the ImageJ software (National Institutes of Health, Bethesda, MD). The protein expressions were reflected by the ratio of the gray value of the target band to that of β-actin band. The protein synthesis inhibitor CHX (SC0353) and proteasome inhibitor MG132 (SC0353) were purchased from Beyotime. The mouse primary cortical neurons were treated with shNC or shRNF8 for 48 h, and then with CHX (50 μg/mL). Cells were collected at 0, 3, 6, and 9 h after administration and HDAC2 protein expression was detected.
Co-IP Detection of HDAC2 Protein Ubiquitination
Mouse primary cortical neurons were transfected with vector, HA-Ub, or Flag-RNF8 plasmids for 36 h, and then treated with 10 μM MG132 for 8 h. Cells were collected and lysed with RIPA lysis buffer (P0013C, Beyotime). Next, 100 μL of cell lysate was incubated with anti-HDAC2 antibody (ab32117, 1:60, Abcam) for 2 h at 4 °C and with 10 μL Protein A Agarose beads overnight. The protein on the beads was extracted, and the ubiquitination level of Ub was detected using anti-HA antibody. At the same time, anti-Flag antibody (ab205606, 1:2000, Abcam) was used to detect the level of Flag and anti-HDAC2 antibody (ab32117, 1:2000, Abcam) was used to detect the level of RNF8 and HDAC2 proteins in cell lysate.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed using EZ-Magna ChIP kits (Millipore). In brief, HEK293T cells at the logarithmic phase of growth were treated with 1% formalin for 1 min to produce DNA–protein cross-linking. Next, the cells were lysed in an ice-water bath and subjected to ultrasonication to produce 200–1000-bp chromatin fragments. Subsequently, 20 μL of the supernatant after centrifugation was taken out as input and the remaining was incubated with 1 μL rabbit antibody to Ac-H3 (07–593, Upstate Biotechnology, Lake Placid, NY) and 1 μL rabbit antibody to IgG (ab172730, Abcam) serving as NC. Each tube was added with 60 μL Protein A Agarose/Salmon Sperm DNA and allowed to stand 10 min, followed by centrifugation at 700 g for 1 min. Subsequently, the precipitate was eluted, after which 20 μL 5 M NaCl was added for de-crosslinking and the DNA was collected. Enriched chromatin fragments were determined by means of RT-qPCR. The sequence of Reelin was forward (F): 5′-CGCGCGCGGGGCCCGTGT-3′ and reverse (R): 5′-AGAGACCGACGGGCTGCC-3′.
Cell Counting Kit-8 (CCK-8) Assay and Lactate Dehydrogenase (LDH) Determination
Proliferation of cells was measured according to the instructions of CCK-8 kit (GK10001, GLPBIO). In brief, 10 μL CCK-8 reagent was added to each well, mixed, and incubated in a cell incubator for 1 h. Next, the optical density values were detected at a wavelength of 450 nm. In addition, the release of LDH in the supernatant of cultured cells was measured using LDH assay kit (C0016, Beyotime) according to the instructions.
Dihydroethidium (DHE) Staining Assay for Detection of Reactive Oxygen Species (ROS) Production in Brain Tissues
The staining solution (ReagentB) was molten in an ice storage tank. The dilution solution (ReagentC) was pre-heated at room temperature. Next, 500 μL ReagentB was added into a 1.5-mL centrifuge tube and mixed with 500 μL ReagentC under conditions void of light. The mouse brain tissues were sliced into 20-μm sections by a freezing microtome and 20 μL pre-cold cleaning solution (ReagentA) was applied to sections. Then, ReagentA was carefully removed and sections were added with ReagentB preheated at room temperature and incubated at 37 °C for 20 min. ReagentB was carefully removed and sections were added with 20 μL ReagentA, which was then removed with care, sealed, and observed under a laser confocal fluorescence microscope with photos taken.
Measurement of Superoxide Dismutase (SOD) and Malondialdehyde (MDA) Production
SOD activity and MDA production were measured in mouse cortical tissues and primary neurons using SOD Activity Assay Kit (ab65354, Abcam) and MDA Assay Kit (ab233471, Abcam), respectively following the manufacturers’ instructions.
Statistical Analysis
All data were processed using GraphPad Prism software. Measurement data were expressed as mean ± standard deviation. Data distribution and homogeneity of variance were first tested. Kolmogorov–Smirnov test was used to verify the normal distribution of the data. p values for all variables greater than 0.05 indicate that the data were normally distributed. Next, Levene’s test was used to test the homogeneity of variance of the data, and the p values greater than 0.05 indicate that the data obey the homogeneity of variance. Data meeting these two criteria were analyzed using parametric tests [27, 28]. Data between two groups were analyzed using unpaired t-test, while multi-group data comparisons were performed by one-way analysis of variance (ANOVA), with Tukey’s post hoc test. In all statistical analysis, a value of p < 0.05 was regarded as statistically significant.
Results
RNF8 Is Poorly Expressed in the Brain Tissues of MCAO Mice and OGD/R-Exposed Neurons
E3 ubiquitin ligase plays an important role in ischemic stroke [9, 29]. In order to explore the possible involvement of E3 ubiquitin ligases in ischemic stroke, we firstly performed differential analysis of the ischemic stroke-related datasets GSE23160, GSE58720, and GSE102541 retrieved from the GEO database and the obtained differentially expressed genes were then intersected. A total of 30 genes were identified, of which, only RNF8 is an E3 ubiquitin ligase (Fig. S1A). Literature has reported that RNF8 deletion leads to neuronal DNA damage and apoptosis, but its role in ischemic stroke, which is pathologically associated with inflammation, oxidative stress, and neuronal apoptosis, has not been fully elucidated [12]. RNF8 was found to be poorly expressed in the ischemic stroke samples in the three datasets (Fig. S1B).
To verify the said results, MCAO mouse models were established using WT mice, which presented with increased infarct volumes compared to the sham-operated mice, indicating successful establishment of the MCAO models (Fig. S1C). In addition, RT-qPCR and western blot analysis results showed that the expression of RNF8 was decreased in the brain tissues of MCAO mice and OGD/R-exposed primary neurons (Fig. S1D, E, Fig. S2A, B).
These findings indicated that RNF8 was poorly expressed in the brain tissues of MCAO mice and OGD/R-exposed neurons.
RNF8 Deficiency Enhances Neuronal Apoptosis and Oxidative Stress in MCAO Mice
To further elucidate the role of RNF8 in ischemic stroke, we first established RNF8−/− MCAO mouse models (Fig. 1A, Fig. S2C). Results from TTC staining illustrated an increase of infarct volume in RNF8−/− MCAO mice (Fig. 1B). At the same time, the neurological deficit scores of RNF8−/− MCAO mice were found to be increased (Fig. 1C). Meanwhile, HE and Nissl staining assays demonstrated depicted a loose arrangement of brain tissues in RNF8−/− MCAO mice, and that many cells in the penumbra exhibited atrophy and pyknosis, along with a decrease in the number of intact neurons (Fig. 1D, E). Moreover, a great number of TUNEL-positive cells were observed in the brain tissues of RNF8−/− MCAO mice (Fig. S3). These results indicated that RNF8 may confer neuroprotective effects against MCAO-induced injury. In addition, ROS production was found to be enhanced in the brain tissues of RNF8−/− MCAO mice (Fig. 1F), while SOD activity was decreased and MDA production was augmented in brain tissues of RNF8−/− MCAO mice (Fig. 1G), suggesting that RNF8 deficiency may promote oxidative stress induced by MCAO. Collectively, these results suggested that RNF8 deficiency could contribute to neuronal apoptosis and oxidative stress in MCAO mice.
RNF8 deficiency leads to neuronal apoptosis and oxidative stress in MCAO mice. A Western blot analysis of RNF8 protein in brain tissues of WT and RNF8−/− mice following MCAO. B TTC staining of brain tissues of WT and RNF8−/− mice following MCAO. C Neurological deficit score of WT and RNF8−/− mice following MCAO. D HE staining of brain tissues of WT and RNF8−/− mice following MCAO. E Nissl staining of brain tissues of WT and RNF8−/− mice following MCAO. F ROS production measured in brain tissues of WT and RNF8−/− mice following MCAO. G SOD activity and MDA production in brain tissues of WT and RNF8−/.− mice following MCAO. *p < 0.05. n = 8 for mice upon each treatment. Data are shown as mean ± standard deviation. Data between two groups were analyzed using unpaired t-test
RNF8 Enhances Ubiquitination and Degradation of HDAC2 in OGD/R-Exposed Neurons
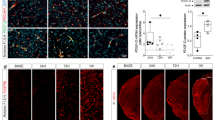
Prediction results from the UbiBrowser indicated that HDAC2 can be used as the substrate of RNF8 (Fig. 2A). Therefore, we speculated that RNF8 may alleviate neuronal apoptosis and oxidative stress induced by MCAO by mediating ubiquitination and degradation of HDAC2. To validate this hypothesis, immunofluorescence was first applied to determine the expression patterns of HDAC2 in the hippocampus of MCAO mice, which revealed an elevation in HDAC2 expression, while the fluorescence of HDAC2 was enhanced in RNF8−/− mice following MCAO compared with WT mice (Fig. 2B).
RNF8 induces HDAC2 ubiquitination and degradation in OGD/R-exposed neurons. A E3 ubiquitin ligase of HDAC2 predicted by the UbiBrowser database. B HDAC2 expression in the brain tissues of WT or RNF8−/.− MCAO mice determined by immunofluorescence. C RNF8 mRNA expression determined by RT-qPCR in OGD/R-exposed neurons treated with shRNF8-1, shRNF8-2, or RNF8 over-expression vector. D Western blot analysis of expression of RNF8 and HDAC2 proteins in response to OGD/R alone or combined with RNF8. E Co-IP analysis of the interaction between RNF8 and HDAC2 in mouse cortical neurons. F Co-IP analysis of the ubiquitination level of HDAC2 in OGD/R-exposed neurons co-treated with RNF8 and MG132 and western blot analysis of HDAC2 and Flag-RNF8 proteins in the lysate of OGD/R-exposed neurons co-treated with RNF8 and MG132. G Western blot analysis of expression of HDAC2 protein in OGD/R-exposed neurons co-treated with shRNF8 and CHX. H Western blot analysis of RNF8 and HDAC2 proteins in OGD/R-exposed neurons treated with RNF8 and MG132 alone or in combination (The OGD/R-exposed neurons were treated with vector or RNF8 for 36 h, and then with 10 μM MG132 for 8 h.). *p < 0.05. n = 8 for mice upon each treatment. Data are shown as mean ± standard deviation. Data between two groups were analyzed using unpaired t-test while multi-group data comparisons were performed by one-way ANOVA with Tukey’s post hoc test. Cell experiments were repeated three times independently
Next, RNF8 was over-expressed or knocked down in mouse cortical neurons, and RT-qPCR confirmed the expression modulation efficiency. Among them, shRNF8-1 exhibited superior silencing efficiency compared to shRNF8-2, which was thus selected for subsequent experiments (Fig. 2C). OGD/R-exposed neurons presented with a decrease in RNF8 protein expression and an increase in HDAC2 protein expression, whereas these effects were reversed following RNF8 over-expression vector treatment (Fig. 2D, Fig. S2D). These results suggested that RNF8 negatively regulated the HDAC2 expression in OGD/R-exposed neurons.
Moreover, the results of Co-IP analysis displayed that RNF8 and HDAC2 could interact with each other in mouse cortical neurons (Fig. 2E). Further results from Co-IP showed that treatment with MG132 promoted the protein expression and ubiquitination levels of HDAC2 (Fig. 2F), demonstrating that RNF8 can promote the ubiquitination level of HDAC2. Meanwhile, silencing of RNF8 prolonged the half-life of HDAC2 as revealed by Chase assay for detection of protein stability (Fig. 2G, Fig. S2E). Upon the addition of MG132, it was found that inhibition of RNF8 on HDAC2 protein expression was significantly abolished (Fig. 2H, Fig. S2F), showing that MG132 could significantly inhibit the degradation of HDAC2 protein by RNF8.
Taken together, these findings indicated that RNF8 interacted with HDAC2 to promote ubiquitination and degradation of HDAC2 in vitro.
RNF8 Attenuates OGD/R-Induced Apoptosis and Oxidative Stress in Cortical Neurons via HDAC2 Inhibition
Next, we sought to illustrate the role of RNF8/HDAC2 axis in apoptosis and oxidative stress of cortical neurons. The results of western blot analysis showed higher expression of RNF8 in cells with RNF8 over-expression vector or in combination with HDAC2 over-expression vector. In addition, HDAC2 expression was found to be reduced upon RNF8 treatment, while it was increased following additional treatment with HDAC2 over-expression vector (Fig. 3A). As shown in Fig. 3B and C, OGD/R exposure decreased the cell viability while increasing the LDH production, and these results were reversed following RNF8 treatment. However, simultaneous over-expression of RNF8 and HDAC2 resulted in a reduction in cell viability and an increase in LDH production. Additionally, TUNEL-positive cells were found to be increased following OGD/R exposure, but they were decreased in response to additional RNF8 treatment. Concomitant over-expression of RNF8 and HDAC2 was observed to increase the number of TUNEL-positive cells (Fig. 3D). These results implied the ability of RNF8 to effectively reverse OGD/R-induced cytotoxicity by inhibiting HDAC2 expression.
RNF8 downregulates HDAC2 expression and thus suppresses the OGD/R-induced apoptosis and oxidative stress in cortical neurons. OGD/R-induced neurons were transduced with RNF8 or in combination with HDAC2. A Western blot analysis of RNF8 and HDAC2 proteins in OGD/R-exposed neurons. B Cell viability of OGD/R-exposed neurons measured by CCK-8 assay. C LDH production measurement in OGD/R-exposed neurons. D TUNEL-positive OGD/R-exposed neurons. E ROS production measurement in OGD/R-exposed neurons. F SOD activity and MDA production in OGD/R-exposed neurons. *p < 0.05. Data are shown as mean ± standard deviation. Multi-group data comparisons were performed by one-way ANOVA with Tukey’s post hoc test. Cell experiments were repeated three times independently
Subsequent results revealed an increase in the fluorescence intensity of DHE upon OGD/R exposure, which was negated following RNF8 treatment, while additional treatment with HDAC2 augmented the fluorescence intensity of DHE (Fig. 3E). In addition, diminished SOD activity and increased MDA production were observed following OGD/R exposure while additional RNF8 treatment counterweighed the results. Moreover, SOD activity was found to be reduced and MDA production was elevated in the presence of concomitant over-expression of RNF8 and HDAC2 (Fig. 3F).
The aforementioned data supported that RNF8 could arrest OGD/R-induced neuronal apoptosis and oxidative stress by inhibiting HDAC2 expression.
HDAC2 Enhances OGD/R-Induced Neuronal Apoptosis and Oxidative Stress via Regulation of the Reelin/GSK3β Axis
We then proceeded to elucidate the downstream mechanism of HDAC2 in OGD/R-exposed neurons. The results of RT-qPCR demonstrated that mRNA expression of Reelin was elevated in primary cortical neurons in response to VPA (an inhibitor of HDAC2), and HDAC2 silencing also resulted in upregulated Reelin expression (Fig. 4A).
HDAC2 inhibits GSK3β-Ser9 phosphorylation by inhibiting the expression of Reelin. A Reelin mRNA expression detected by RT-qPCR in OGD/R-exposed neurons treated with VPA or shHDAC2. B Enrichment of H3K27me3 in the Reelin promoter region measured by ChIP assay in OGD/R-exposed neurons treated with VPA or shHDAC2. C Acetylation level of H3K27me3 in the Reelin promoter measured by ChIP assay in OGD/R-exposed neurons treated with shHDAC2. D Reelin mRNA expression detected by RT-qPCR in OGD/R-exposed neurons treated with shHDAC2. E Reelin mRNA expression detected by RT-qPCR in OGD/R-exposed neurons treated with HDAC2 alone or combined with Reelin. F Western blot analysis of HDAC2 and Reelin proteins and GSK3β-Ser9 phosphorylation level in OGD/R-exposed neurons treated with HDAC2 alone or combined with Reelin. *p < 0.05. Data are shown as mean ± standard deviation. Data between two groups were analyzed using unpaired t-test, while multi-group data comparisons were performed by one-way ANOVA with Tukey’s post hoc test. Cell experiments were repeated three times independently
Meanwhile, the results of ChIP assay demonstrated that VPA treatment or HDAC2 silencing could promote the enrichment of H3K27me3 in the Reelin promoter region (Fig. 4B). As shown in Fig. 4C and D, OGD/R exposure increased the acetylation of H3K27me3 in the Reelin promoter and decreased the mRNA expression of Reelin but further knockdown of HDAC2 abrogated this effect. These results suggested that HDAC2 inhibited the transcription of Reelin by inhibiting the acetylation of H3K27me3.
Additionally, the results of RT-qPCR and western blot analysis revealed a decline in the Reelin protein expression and GSK3β-Ser9 phosphorylation levels after OGD/R exposure, while opposite effects were noted upon simultaneous over-expression of HDAC2 and Reelin. In addition, HDAC2 downregulated the expression of Reelin and GSK3β-Ser9 phosphorylation levels (Fig. 4E, F). These data indicated that HDAC2 inhibited GSK3β-Ser9 phosphorylation by inhibiting the expression of Reelin.
Furthermore, Fig. 5A and B illustrate that the diminished cell viability and elevated LDH production in HDAC2-treated OGD/R cells could be abrogated following simultaneous over-expression of HDAC2 and Reelin. In addition, the promoting effect of HDAC2 on TUNEL-positive cells was noted to be reversed upon both HDAC2 and Reelin treatment (Fig. 5C). These findings uncovered that HDAC2 facilitated OGD/R-induced cytotoxicity by inhibiting the Reelin expression. Furthermore, an increase was noted in the fluorescence intensity of DHE in OGD/R cells over-expressing HDAC2, while a reduction was observed in the presence of concomitant over-expression of HDAC2 and Reelin (Fig. 5D). Meanwhile, decreased SOD activity and enhanced MDA production were noted in cells over-expressing HDAC2, the effect of which could be abolished by simultaneous over-expression of HDAC2 and Reelin (Fig. 5E).
HDAC2 potentiates OGD/R-induced apoptosis and oxidative stress via the Reelin/GSK3β axis. A Cell viability in OGD/R-exposed neurons upon treatment with HDAC2 alone or combined with Reelin measured by CCK-8 assay. B LDH production measurement in OGD/R-exposed neurons treated with HDAC2 alone or combined with Reelin. C TUNEL-positive OGD/R-exposed neurons treated with HDAC2 alone or combined with Reelin. D ROS production measurement in OGD/R-exposed neurons treated with HDAC2 alone or combined with Reelin. E SOD activity and MDA production in OGD/R-exposed neurons treated with HDAC2 alone or combined with Reelin. *p < 0.05. Data are shown as mean ± standard deviation. Multi-group data comparisons were performed by one-way ANOVA with Tukey’s post hoc test. Cell experiments were repeated three times independently
Altogether, these findings indicated that HDAC2 facilitated the OGD/R-induced apoptosis and oxidative stress by regulating the Reelin/GSK3β axis.
RNF8 Alleviates Ischemic Stroke Through Regulation of the Reelin/GSK3β Axis In Vivo
In view of the above-mentioned experimental results, we speculated that RNF8 may regulate the Reelin/GSK3β axis to alleviate ischemic stroke. Analysis of expression data in the brain tissue samples of MCAO mice in the GSE23160 dataset together with Pearson’s correlation coefficient revealed that the expression of RNF8 was positively correlated with the Reelin expression in MCAO mouse brain tissues (Fig. 6A). Moreover, RT-qPCR results showed a reduction in the Reelin expression in brain tissues of MCAO mice or RNF8−/− mice following MCAO induction (Fig. 6B). Meanwhile, western blot analysis results showed that Reelin expression as well as GSK3β-Ser9 phosphorylation levels was augmented in the brain tissues of Reelin-treated WT mice or RNF8−/− MCAO mice, whereas these levels were found to be decreased following treatment with RNF8−/− alone in MCAO mice (Fig. 6C, Fig. S2G). In addition, infarct volume and neurological deficit scores exhibited a decline in WT or RNF8−/− MCAO mice treated with Reelin (Fig. 6D, E).
RNF8 exerts neuroprotective effects against ischemic stroke via regulation of the Reelin/GSK3β axis in vivo. A Correlation of RNF8 expression with Reelin expression in the brain tissue samples of MCAO mice (n = 24) based on the expression data in the GSE23160 dataset analyzed by Pearson’s correlation coefficient (r2 = 0.581, p < 0.01). B Reelin mRNA expression in brain tissues of untreated WT or RNF8−/− mice or MCAO-induced WT or RNF8−/− mice determined by RT-qPCR. C Western blot analysis of RNF8, GSK3β, HDAC2, and Reelin proteins along with GSK3β-Ser9 phosphorylation level in mouse brain tissues of WT or RNF8−/− MCAO mice with/without Reelin treatment. D TTC staining of mouse brain tissues of WT or RNF8−/− MCAO mice with/without Reelin treatment. E Neurological deficit scores of WT or RNF8−/− MCAO mice with/without Reelin treatment. F HE staining of mouse brain tissues of WT or RNF8−/− MCAO mice with/without Reelin treatment. G Nissl staining of mouse brain tissues of WT or RNF8−/− MCAO mice with/without Reelin treatment. H TUNEL-positive cells in mouse brain tissues of WT or RNF8−/− MCAO mice with/without Reelin treatment. I ROS production in mouse brain tissues of WT or RNF8−/− MCAO mice with/without Reelin treatment measured by DHE staining. J SOD activity and MDA production in mouse brain tissues of WT or RNF8.−/− MCAO mice with/without Reelin treatment. *p < 0.05. n = 8 for mice upon each treatment. Data are shown as mean ± standard deviation. Multi-group data comparisons were performed by one-way ANOVA with Tukey’s post hoc test
Furthermore, morphological observation of neurons of WT or RNF8−/− mice following MCAO induction using HE and Nissl staining assays displayed more compact brain tissues, complete and clear nuclei of cells in penumbra, and greater number of complete neurons in response to additional Reelin treatment (Fig. 6F, G). The TUNEL-positive cells were observed to be decreased in brain tissues of WT or RNF8−/− mice following MCAO induction in the presence of Reelin (Fig. 6H). These results indicated that RNF8 had neuroprotective effects against MCAO-induced injury through Reelin upregulation. Additionally, in brain tissues of WT or RNF8−/− mice following MCAO induction treated with Reelin, ROS and MDA production was decreased while SOD activity was augmented (Fig. 6I, J), suggesting that RNF8 hindered the MCAO-induced oxidative stress through upregulation of Reelin.
In summary, the above data indicated that RNF8 repressed MCAO-induced apoptosis and oxidative stress and thus delayed ischemic stroke through regulation of the Reelin/GSK3β axis in vivo.
Discussion
Ischemic stroke is one of the leading causes of neuronal injury or death and can eventually lead to severe neurological disorders or even death in certain circumstances [30]. The current study indicated that RNF8 could potentially confer a significant neuroprotective role against ischemic stroke by downregulating HDAC2 and eliciting Reelin-mediated GSK3β inhibition, thus alleviating ischemic stroke.
The experimental data in our study demonstrated that RNF8 was poorly expressed in the brain tissues of MCAO mice and in OGD/R-exposed neurons. Moreover, we further uncovered that deficiency in RNF8 could lead to neuronal damage, apoptosis, and oxidative stress in MCAO mice, as evidenced by increased TUNEL-positive cells, enhanced ROS and MDA production, yet decreased SOD activity. Meanwhile, a previous study presented evidence suggesting that RNF8 deficiency enhances DNA damage accumulation, neuronal apoptosis, neuronal degeneration, and reactive astrocytosis [12]. The attenuated oxidative stress is accompanied by increased SOD activity and reduced ROS and MDA production [31].
Another significant finding made in our study was that RNF8 could enhance the ubiquitination and degradation of HDAC2. Results from the UbiBrowser database indicated HDAC2 as the substrate of RNF8, which was found to negatively regulate the expression of HDAC2 in stroke. In accordance with our findings, previous evidence also documented that targeting the ubiquitination and degradation of HDAC2 could trigger the reduction of its expression [32, 33]. Meanwhile, HDAC2 was upregulated following ischemia stroke, whereas its suppression in the peri-infarct cortex of rodents could promote recovery of neurological deficit from stroke by epigenetically enhancing cell survival and neuroplasticity of surviving neurons as well as reducing neuroinflammation [34]. Moreover, inhibition of HDAC2 conferred a neuroprotective role, attributed to the reduction in infarct volume and neuronal apoptosis in the cerebral cortex of the damaged hemisphere in photothrombotic stroke mice [35]. More importantly, blockage of HDAC2 activity via microRNA-455-3p exhibits significant correlation with suppression of oxidative stress [36]. Consequently, it would plausible to suggest that RNF8 may attenuate neuronal apoptosis and oxidative stress following ischemic stroke via HDAC2 inhibition.
Furthermore, deficiency in Reelin resulted in attenuation of proliferation and migration of the cerebellar neurons [37]. On the other hand, upregulation of Reelin and reduced oxidative stress is known to protect against neurodegeneration and improve brain plasticity [38]. Mechanistic investigations in our study showed that HDAC2 inhibited Reelin expression and resulted in a reduction in GSK3β-Ser9 phosphorylation. Prior evidence has suggested that Reelin-deficient mice subjected to Alzheimer’s disease presented with decreased GSK3β phosphorylation, while GSK3β activity was enhanced [19]. GSK3β is known to function as an inhibitor to downregulate Nrf2 expression, and thus disrupts its protective role against oxidative stress in brain ischemia and reperfusion injury [39]. Besides, increased GSK3β phosphorylation by lemur tyrosine kinase-2 can facilitate the alleviation of OGD/R-induced neuronal apoptosis and oxidative damage [40]. The aforementioned data helped us infer that HDAC2 augments OGD/R-induced apoptosis and oxidative stress via regulation of the Reelin/GSK3β axis. More importantly, an inverse correlation between RNF8 and GSK3β was previously established during the progression of breast cancer [41], whereas RNF8 and Reelin are known to be positively correlated according to the analysis results of the GSE23160 dataset. Overall, our findings in conjunction with existing data indicate that RNF8 upregulates the Reelin expression and induces the resultant GSK3β inhibition, contributing to the alleviation of ischemic stroke.
Altogether, our study demonstrates that RNF8 holds potential neuroprotective properties against ischemic stroke through the HDAC2/Reelin/GSK3β axis (Fig. 7). Our data may provide novel insights in the development of new therapies for the alleviation of ischemic stroke. However, the expression of RNF8 in different brain regions (cortex, striatum, thalamus, hippocampus, and subventricular regions of sham-operated and MCAO mice) should be measured. Moreover, further investigation warrants for the effect of the pharmacological modulation of RNF8 (inhibition and activation) on the observed neuroprotective effect. In addition, whether the therapeutic target is applicable to human beings needs to be further explored. Besides, the findings obtained in this study are preliminary, indicating more studies in this area are required in the future.
Data Availability
The data and materials of the study can be obtained from the corresponding author upon request.
Change history
05 July 2022
A Correction to this paper has been published: https://doi.org/10.1007/s12035-022-02938-9
Abbreviations
- RNF8:
-
RING finger protein 8
- MCAO:
-
Middle cerebral artery occlusion
- OGD/R:
-
Oxygen-glucose deprivation/reoxygenation
- HDAC2:
-
Histone deacetylase 2
- GSK3β:
-
Glycogen synthase kinase-3beta
- HDACs:
-
Histone deacetylases
- CBF:
-
Cerebral blood flow
- PCR:
-
Polymerase chain reaction
- TTC:
-
Triphenyl tetrazolium chloride
- HE:
-
Hematoxylin-eosin
- PBS:
-
Phosphate buffer saline
- shRNA:
-
Short hairpin RNA
- Co-IP:
-
Co-immunoprecipitation
- TBS:
-
Tris-buffered saline
- TUNEL:
-
Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling
- ChIP:
-
Chromatin immunoprecipitation
- CCK-8:
-
Cell counting kit-8
- LDH:
-
Lactate dehydrogenase
- DHE:
-
Dihydroethidium
- ROS:
-
Reactive oxygen species
- SOD:
-
Superoxide dismutase
- MDA:
-
Malondialdehyde
References
Campbell BCV, Khatri P (2020) Stroke Lancet 396(10244):129–142
Yaghi S, Ishida K, Torres J, Mac Grory B, Raz E, Humbert K et al (2020) SARS-CoV-2 and stroke in a New York healthcare system. Stroke 51(7):2002–2011
Wang Z, Yang Y, Liang X, Gao B, Liu M, Li W et al (2020) COVID-19 associated ischemic stroke and hemorrhagic stroke: incidence, potential pathological mechanism, and management. Front Neurol 11:571996
Smith M, Reddy U, Robba C, Sharma D, Citerio G (2019) Acute ischaemic stroke: challenges for the intensivist. Intensive Care Med 45(9):1177–1189
Weaver NA, Kuijf HJ, Aben HP, Abrigo J, Bae HJ, Barbay M et al (2021) Strategic infarct locations for post-stroke cognitive impairment: a pooled analysis of individual patient data from 12 acute ischaemic stroke cohorts. Lancet Neurol 20(6):448–459
Saber H, Thrift AG, Kapral MK, Shoamanesh A, Amiri A, Farzadfard MT et al (2017) Incidence, recurrence, and long-term survival of ischemic stroke subtypes: a population-based study in the Middle East. Int J Stroke 12(8):835–843
Kessner SS, Schlemm E, Cheng B, Bingel U, Fiehler J, Gerloff C et al (2019) Somatosensory deficits after ischemic stroke. Stroke 50(5):1116–1123
Arumugam TV, Baik SH, Balaganapathy P, Sobey CG, Mattson MP, Jo DG (2018) Notch signaling and neuronal death in stroke. Prog Neurobiol 165–167:103–116
Kim T, Chokkalla AK, Vemuganti R (2020) Deletion of ubiquitin ligase Nedd4l exacerbates ischemic brain damage. J Cereb Blood Flow Metab:271678X20943804
Li X, Liao Y, Dong Y, Li S, Wang F, Wu R et al (2020) Mib2 deficiency inhibits microglial activation and alleviates ischemia-induced brain injury. Aging Dis 11(3):523–535
Kuang J, Min L, Liu C, Chen S, Gao C, Ma J et al (2020) RNF8 promotes epithelial-mesenchymal transition in lung cancer cells via stabilization of slug. Mol Cancer Res 18(11):1638–1649
Ouyang S, Song Y, Tian Y, Chen Y, Yu X, Wang D (2015) RNF8 deficiency results in neurodegeneration in mice. Neurobiol Aging 36(10):2850–2860
Seto E, Yoshida M (2014) Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6(4):a018713
Falkenberg KJ, Johnstone RW (2014) Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 13(9):673–691
Demyanenko SV, Dzreyan VA, Neginskaya MA, Uzdensky AB (2020) Expression of histone deacetylases HDAC1 and HDAC2 and their role in apoptosis in the penumbra induced by photothrombotic stroke. Mol Neurobiol 57(1):226–238
Sato N, Fukushima N, Chang R, Matsubayashi H, Goggins M (2006) Differential and epigenetic gene expression profiling identifies frequent disruption of the RELN pathway in pancreatic cancers. Gastroenterology 130(2):548–565
Ampuero E, Jury N, Hartel S, Marzolo MP, van Zundert B (2017) Interfering of the Reelin/ApoER2/PSD95 signaling axis reactivates dendritogenesis of mature hippocampal neurons. J Cell Physiol 232(5):1187–1199
Stary CM, Xu L, Sun X, Ouyang YB, White RE, Leong J et al (2015) MicroRNA-200c contributes to injury from transient focal cerebral ischemia by targeting Reelin. Stroke 46(2):551–556
Ohkubo N, Lee YD, Morishima A, Terashima T, Kikkawa S, Tohyama M et al (2003) Apolipoprotein E and Reelin ligands modulate tau phosphorylation through an apolipoprotein E receptor/disabled-1/glycogen synthase kinase-3beta cascade. FASEB J 17(2):295–297
Chen C, Yu Q, Xu K, Cai L, Felicia BM, Wang L et al (2020) Electroacupuncture pretreatment prevents ischemic stroke and inhibits Wnt signaling-mediated autophagy through the regulation of GSK-3beta phosphorylation. Brain Res Bull 158:90–98
Wang Y, Meng C, Zhang J, Wu J, Zhao J (2019) Inhibition of GSK-3beta alleviates cerebral ischemia/reperfusion injury in rats by suppressing NLRP3 inflammasome activation through autophagy. Int Immunopharmacol 68:234–241
Modzelewski AJ, Hilz S, Crate EA, Schweidenback CT, Fogarty EA, Grenier JK et al (2015) Dgcr8 and Dicer are essential for sex chromosome integrity during meiosis in males. J Cell Sci 128(12):2314–2327
Valnegri P, Huang J, Yamada T, Yang Y, Mejia LA, Cho HY et al (2017) RNF8/UBC13 ubiquitin signaling suppresses synapse formation in the mammalian brain. Nat Commun 8(1):1271
Wang S, Mou J, Cui L, Wang X, Zhang Z (2018) Astragaloside IV inhibits cell proliferation of colorectal cancer cell lines through down-regulation of B7–H3. Biomed Pharmacother 102:1037–1044
Zhao Y, Majid MC, Soll JM, Brickner JR, Dango S, Mosammaparast N (2015) Noncanonical regulation of alkylation damage resistance by the OTUD4 deubiquitinase. EMBO J 34(12):1687–1703
Sun F, Zhang Y, Xu L, Li S, Chen X, Zhang L et al (2018) Proteasome inhibitor MG132 enhances cisplatin-induced apoptosis in osteosarcoma cells and inhibits tumor growth. Oncol Res 26(4):655–664
Wu J, He J, Tian X, Li H, Wen Y, Shao Q et al (2020) Upregulation of miRNA-9-5p promotes angiogenesis after traumatic brain injury by inhibiting Ptch-1. Neuroscience 440:160–174
Jiang Q, Geng X, Warren J, Eugene Paul Cosky E, Kaura S, Stone C et al (2020) Hypoxia inducible factor-1alpha (HIF-1alpha) mediates NLRP3 inflammasome-dependent-pyroptotic and apoptotic cell death following ischemic stroke. Neuroscience 448:126–139
Li T, Qin JJ, Yang X, Ji YX, Guo F, Cheng WL et al (2017) The ubiquitin E3 ligase TRAF6 exacerbates ischemic stroke by ubiquitinating and activating Rac1. J Neurosci 37(50):12123–12140
Chen YM, He XZ, Wang SM, Xia Y (2020) Delta-opioid receptors, microRNAs, and neuroinflammation in cerebral ischemia/hypoxia. Front Immunol 11:421
Wang L, Shi Z, Wang X, Mu S, Xu X, Shen L et al (2021) Protective effects of bovine milk exosomes against oxidative stress in IEC-6 cells. Eur J Nutr 60(1):317–327
Zhang J, Kan S, Huang B, Hao Z, Mak TW, Zhong Q (2011) Mule determines the apoptotic response to HDAC inhibitors by targeted ubiquitination and destruction of HDAC2. Genes Dev 25(24):2610–2618
Yao H, Rahman I (2012) Role of histone deacetylase 2 in epigenetics and cellular senescence: implications in lung inflammaging and COPD. Am J Physiol Lung Cell Mol Physiol 303(7):L557-566
Lin YH, Dong J, Tang Y, Ni HY, Zhang Y, Su P et al (2017) Opening a new time window for treatment of stroke by targeting HDAC2. J Neurosci 37(28):6712–6728
Demyanenko SV, Nikul VV, Uzdensky AB (2020) The neuroprotective effect of the HDAC2/3 inhibitor MI192 on the penumbra after photothrombotic stroke in the mouse brain. Mol Neurobiol 57(1):239–248
Zhang S, Wu W, Jiao G, Li C, Liu H (2018) MiR-455-3p activates Nrf2/ARE signaling via HDAC2 and protects osteoblasts from oxidative stress. Int J Biol Macromol 107(Pt B):2094–2101
Cocito C, Merighi A, Giacobini M, Lossi L (2016) Alterations of cell proliferation and apoptosis in the hypoplastic reeler cerebellum. Front Cell Neurosci 10:141
Herring A, Donath A, Yarmolenko M, Uslar E, Conzen C, Kanakis D et al (2012) Exercise during pregnancy mitigates Alzheimer-like pathology in mouse offspring. FASEB J 26(1):117–128
Chen X, Liu Y, Zhu J, Lei S, Dong Y, Li L et al (2016) GSK-3beta downregulates Nrf2 in cultured cortical neurons and in a rat model of cerebral ischemia-reperfusion. Sci Rep 6:20196
Bao H, Gao M (2020) Overexpression of lemur tyrosine kinase-2 protects neurons from oxygen-glucose deprivation/reoxygenation-induced injury through reinforcement of Nrf2 signaling by modulating GSK-3beta phosphorylation. Biochem Biophys Res Commun 521(4):964–970
Kuang J, Li L, Guo L, Su Y, Wang Y, Xu Y et al (2016) RNF8 promotes epithelial-mesenchymal transition of breast cancer cells. J Exp Clin Cancer Res 35(1):88
Funding
This study was supported by National Natural Science Foundation (81860321); Science and Technology Planning Project of Baiyun District, Guiyang City, Baike Contract [(2019) No. 36]; Regional Common Diseases and Adult Stem Cell Transformation Research Innovation Platform, Science and Technology Department of Guizhou Province [Guizhou Specific Grant (2019) 4008]; The National Natural Science Foundation of China (Project No. 82060232); Science and Technology Plan Project of Guizhou Province, Science and Technology Department of Guizhou Province [Basic Science and Technology Cooperation [(2020) 1Z060]]; Guizhou Science Foundation [(2017) 1149]; and Guizhou Province Science and Technology Planning Project [Guizhou Genetic Basis-ZK [2021] General 489].
Author information
Authors and Affiliations
Contributions
Zhi Huang and Xiaoxi Zhu designed the study, collated the data, designed and developed the database, carried out data analyses, and produced the initial draft of the manuscript. Zhi Huang and Xiaoxi Zhu contributed to drafting the manuscript. Junxiang Li, Dengwei You, Yan Xiao, and Wenfeng Yu edited and revised the manuscript. All authors have read and approved the final submitted manuscript.
Corresponding authors
Ethics declarations
Ethics Approval
The current study was carried out under the approval of the Ethics committee of the Affiliated Hospital of Guizhou Medical University (approval number: 2020-046A), and conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
Consent to Participate
Not applicable.
Consent for Publication
Consent for publication was obtained from the participants.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaoxi Zhu, Junxiang Li, and Dengwei You are regarded as co-first authors.
Supplementary Information
Below is the link to the electronic supplementary material.



Rights and permissions
About this article
Cite this article
Zhu, X., Li, J., You, D. et al. Neuroprotective Effect of E3 Ubiquitin Ligase RNF8 Against Ischemic Stroke via HDAC2 Stability Reduction and Reelin-Dependent GSK3β Inhibition. Mol Neurobiol 59, 4776–4790 (2022). https://doi.org/10.1007/s12035-022-02880-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-02880-w