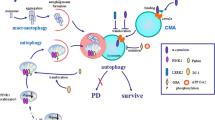
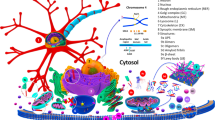
Abstract
Over the last decade, researchers have discovered that a group of apparently unrelated neurodegenerative disorders, such as Parkinson's disease, have remarkable cellular and molecular biology similarities. Protein misfolding and aggregation are involved in all of the neurodegenerative conditions; as a result, inclusion bodies aggregation starts in the cells. Chaperone proteins and ubiquitin (26S proteasome's proteolysis signal), which aid in refolding misfolded proteins, are frequently found in these aggregates. The discovery of disease-causing gene alterations that code for multiple ubiquitin–proteasome pathway proteins in Parkinson's disease has strengthened the relationship between the ubiquitin–proteasome system and neurodegeneration. The specific molecular linkages between these systems and pathogenesis, on the other hand, are unknown and controversial. We outline the current level of knowledge in this article, focusing on important unanswered problems.
Graphical Abstract





Similar content being viewed by others
Data Availability
Not applicable.
Abbreviations
- AA:
-
amino acid
- AD:
-
Alzheimer's disease
- ATG:
-
autophagy-related gene
- CDCrel-1:
-
cell division control-related protein
- CMA:
-
chaperone-mediated autophagy
- CP:
-
core particle
- E1:
-
ubiquitin-activating enzyme
- E2:
-
ubiquitin-conjugating enzyme
- E3:
-
ubiquitin-ligating enzyme
- GstS1:
-
glutathione S-transferase S1
- GABARAP:
-
g-aminobutyric acid receptor-associated protein
- HSP 90:
-
heat shock protein 90
- KO:
-
knock out
- LC:
-
locus coeruleus
- LC3:
-
microtubule-associated protein 1 light chain 3
- Pael-R:
-
parkin-associated endothelial-like receptor
- PD:
-
Parkinson's disease
- PROTAC:
-
proteolysis targeting chimera
- Sfp1:
-
split-finger protein 1
- SnPc:
-
substantia nigra pars compacta
- Ub:
-
ubiquitin
- UCH-L1:
-
carboxyl-terminal hydrolase L1
- UPS:
-
ubiquitin–proteasome system
References
Wijeyekoon RS, Moore SF, Farrell K, Breen DP, Barker RA, Williams-Gray CH (2020) Cerebrospinal Fluid Cytokines and Neurodegeneration-Associated Proteins in Parkinson's Disease. Mov Disord 35:1062–1066. https://doi.org/10.1002/mds.28015
Nalls MA, Duran R, Lopez G, Kurzawa-Akanbi M, McKeith IG, Chinnery PF, Morris CM, Theuns J, Crosiers D, Cras P, Engelborghs S (2013) A multicenter study of glucocerebrosidase mutations in dementia with Lewy bodies. JAMA Neurol 70:727–735. https://doi.org/10.1001/jamaneurol.2013.1925
Guerreiro R, Escott-Price V, Darwent L, Parkkinen L, Ansorge O, Hernandez DG, Nalls MA, Clark L, Honig L, Marder K, van der Flier W (2016) Genome-wide analysis of genetic correlation in dementia with Lewy bodies, Parkinson's and Alzheimer's diseases. Neurobiol Aging 38:214–2e7. https://doi.org/10.1016/j.neurobiolaging.2015.10.028
Singh A, Dawson TM, Kulkarni S (2021) Neurodegenerative disorders and gut-brain interactions. J Clin Investig 131:e143775. https://doi.org/10.1172/JCI143775
Kelleher RJ, Shen J (2017) Presenilin-1 mutations and Alzheimer’s disease. Proc Natl Acad Sci 114:629–631. https://doi.org/10.1073/pnas.1619574114
Dawkins E, Small DH (2014) Insights into the physiological function of the β-amyloid precursor protein: beyond Alzheimer's disease. J Neurochem 129:756–769. https://doi.org/10.1111/jnc.12675
Saccon RA, Bunton-Stasyshyn RK, Fisher EM, Fratta P (2013) Is SOD1 loss of function involved in amyotrophic lateral sclerosis? Brain 136:2342–2358. https://doi.org/10.1093/brain/awt097
Sieradzan KA, Mechan AO, Jones L, Wanker EE, Nukina N, Mann DM (1999) Huntington's disease intranuclear inclusions contain truncated, ubiquitinated huntingtin protein. Exp Neurol 156:92–99. https://doi.org/10.1006/exnr.1998.7005
Scoles DR, Meera P, Schneider MD, Paul S, Dansithong W, Figueroa KP, Hung G, Rigo F, Bennett CF, Otis TS, Pulst SM (2017) Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 544:362–366. https://doi.org/10.1038/nature22044
Zhu Q, Zhuang XX, Chen JY, Yuan NN, Chen Y, Cai CZ, Tan JQ, Su HX, Lu JH (2021) Lycorine, a natural alkaloid, promotes the degradation of alpha-synuclein via PKA-mediated UPS activation in transgenic Parkinson's disease models. Phytomedicine 87:153578. https://doi.org/10.1016/j.phymed.2021.153578
Ham SJ, Lee D, Xu WJ, Cho E, Choi S, Min S, Park S, Chung J (2021) Loss of UCHL1 rescues the defects related to Parkinson’s disease by suppressing glycolysis. Sci Adv 7:eabg4574. https://doi.org/10.1126/sciadv.abg4574
Hirsch EC, Standaert DG (2021) Ten unsolved questions about neuroinflammation in Parkinson's disease. Mov Disord 36:16–24. https://doi.org/10.1002/mds.28075
Jiang P, Dickson DW (2018) Parkinson’s disease: experimental models and reality. Acta Neuropathol 135:13–32. https://doi.org/10.1007/s00401-017-1788-5
Mulder MP, Witting KF, Ovaa H (2019) Cracking the ubiquitin Code: the ubiquitin toolbox. Curr Issues Mol Biol 37:1–20. https://doi.org/10.21775/cimb.037.001
Cao Y, Zhu H, He R, Kong L, Shao J, Zhuang R, Xi J, Zhang J (2020) Proteasome, a promising therapeutic target for multiple diseases beyond cancer. Drug Des Devel Ther 14:4327. https://doi.org/10.2147/DDDT.S265793
Kist M, Vucic D (2021) Cell death pathways: intricate connections and disease implications. EMBO J 40:e106700. https://doi.org/10.15252/embj.2020106700
Webster CP, Smith EF, Shaw PJ, De Vos KJ (2017) Protein homeostasis in amyotrophic lateral sclerosis: therapeutic opportunities? Front Mol Neurosci 10:123. https://doi.org/10.3389/fnmol.2017.00123
Kalkan AC, Kahraman T, Ugut BO, Colakoglu BD, Genc A (2020) A comparison of the relationship between manual dexterity and postural control in young and older individuals with Parkinson’s disease. J Clin Neurosci 75:89–93. https://doi.org/10.1016/j.jocn.2020.03.018
Osellame LD, Duchen MR (2013) Defective quality control mechanisms and accumulation of damaged mitochondria link Gaucher and Parkinson diseases. Autophagy 9:1633–1635. https://doi.org/10.4161/auto.25878
Liddell JR, White AR (2018) Nexus between mitochondrial function, iron, copper and glutathione in Parkinson's disease. Neurochem Int 117:126–138. https://doi.org/10.1016/j.neuint.2017.05.016
Vila M, Laguna A, Carballo-Carbajal I (2019) Intracellular crowding by age-dependent neuromelanin accumulation disrupts neuronal proteostasis and triggers Parkinson disease pathology. Autophagy 15:2028–2030. https://doi.org/10.1080/15548627.2019.1659621
McNaught KS, Perl DP, Brownell AL, Olanow CW (2004) Systemic exposure to proteasome inhibitors causes a progressive model of Parkinson's disease. Ann Neurol 56:149–162. https://doi.org/10.1002/ana.20186
Bové J, Zhou C, Jackson-Lewis V, Taylor J, Chu Y, Rideout HJ, Wu DC, Kordower JH, Petrucelli L, Przedborski S (2006) Proteasome inhibition and Parkinson's disease modeling. Ann Neurol 60:260–264. https://doi.org/10.1002/ana.20937
Zeng BY, Bukhatwa S, Hikima A, Rose S, Jenner P (2006) Reproducible nigral cell loss after systemic proteasomal inhibitor administration to rats. Ann Neurol 60:248–252. https://doi.org/10.1002/ana.20932
Bedford L, Hay D, Devoy A, Paine S, Powe DG, Seth R, Gray T, Topham I, Fone K, Rezvani N, Mee M (2008) Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J Neurosci 28:8189–8198. https://doi.org/10.1523/JNEUROSCI.2218-08.2008
Kordower JH, Kanaan NM, Chu Y, Suresh Babu R, Stansell J III, Terpstra BT, Sortwell CE, Steece-Collier K, Collier TJ (2006) Failure of proteasome inhibitor administration to provide a model of Parkinson's disease in rats and monkeys. Ann Neurol 60:264–268. https://doi.org/10.1002/ana.20935
Jiang H, Yu Y, Liu S, Zhu M, Dong X, Wu J, Zhang Z, Zhang M, Zhang Y (2019) Proteomic study of a Parkinson's disease model of undifferentiated SH-SY5Y cells induced by a proteasome inhibitor. Int J Med Sci 16:84. https://doi.org/10.7150/ijms.28595
Bentea E, Verbruggen L, Massie A (2017) The proteasome inhibition model of Parkinson’s disease. J Parkinsons Dis 7:31–63. https://doi.org/10.3233/JPD-160921
Greene ER, Dong KC, Martin A (2020) Understanding the 26S proteasome molecular machine from a structural and conformational dynamics perspective. Curr Opin Struct Biol 61:33–41. https://doi.org/10.1016/j.sbi.2019.10.004
Demasi M, da Cunha FM (2018) The physiological role of the free 20S proteasome in protein degradation: a critical review. BBA 1862:2948–2954. https://doi.org/10.1016/j.bbagen.2018.09.009
Kommaddi RP, Shenoy SK (2013) Arrestins and protein ubiquitination. Prog Mol Biol Transl Sci 118:175–204. https://doi.org/10.1016/B978-0-12-394440-5.00007-3
Wang Y, Tang C, Wang E, Wang J (2014) PolyUbiquitin chain linkage topology selects the functions from the underlying binding landscape. PLoS Comput Biol 10:e1003691. https://doi.org/10.1371/journal.pcbi.1003691
Luza S, Opazo CM, Bousman CA, Pantelis C, Bush AI, Everall IP (2020) The ubiquitin proteasome system and schizophrenia. Lancet Psychiatry 7:528–537. https://doi.org/10.1016/S2215-0366(19)30520-6
Colberg L, Cammann C, Greinacher A, Seifert U (2020) Structure and function of the ubiquitin-proteasome system in platelets. J Thromb Haemost 18:771–780. https://doi.org/10.1111/jth.14730
Mao Y (2021) Structure, dynamics and function of the 26S proteasome. InMacromolecular Protein Complexes III: Structure and Function. Springer, Cham p. 1-151. https://doi.org/10.1007/978-3-030-58971-4_1
Davis C, Spaller BL, Matouschek A (2021) Mechanisms of substrate recognition by the 26S proteasome. Curr Opin Struct 67:161–169. https://doi.org/10.1016/j.sbi.2020.10.010
Butler EK, Voigt A, Lutz AK, Toegel JP, Gerhardt E, Karsten P, Falkenburger B, Reinartz A, Winklhofer KF, Schulz JB (2012) The mitochondrial chaperone protein TRAP1 mitigates α-Synuclein toxicity. PLoS Genet 8:e1002488. https://doi.org/10.1371/journal.pgen.1002488
Klucken J, Shin Y, Masliah E, Hyman BT, McLean PJ (2004) Hsp70 reduces α-synuclein aggregation and toxicity. J Biol Chem 279:25497–25502. https://doi.org/10.1074/jbc.M400255200
Wang XJ, Yu J, Wong SH, Cheng AS, Chan FK, Ng SS, Cho CH, Sung JJ, Wu WK (2013) A novel crosstalk between two major protein degradation systems: regulation of proteasomal activity by autophagy. Autophagy 9:1500–1508. https://doi.org/10.4161/auto.25573
Bao XQ, Kong XC, Qian C, Zhang D (2012) FLZ protects dopaminergic neuron through activating protein kinase B/mammalian target of rapamycin pathway and inhibiting RTP801 expression in Parkinson's disease models. Neuroscience 202:396–404. https://doi.org/10.1016/j.neuroscience.2011.11.036
Sakata E, Eisele MR, Baumeister W (2020) Molecular and cellular dynamics of the 26S proteasome. Biochim Biophys Acta, Proteins Proteomics 13:140583. https://doi.org/10.1016/j.bbapap.2020.140583
Martinez-Fonts K, Davis C, Tomita T, Elsasser S, Nager AR, Shi Y, Finley D, Matouschek A (2020) The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat Commun 11:1–6. https://doi.org/10.1038/s41467-019-13906-8
Budenholzer L, Cheng CL, Li Y, Hochstrasser M (2017) Proteasome structure and assembly. J Mol Biol 429:3500–3524. https://doi.org/10.1016/j.jmb.2017.05.027
Thibaudeau TA, Smith DM (2019) A practical review of proteasome pharmacology. Pharmacol Rev 71:170–197. https://doi.org/10.1124/pr.117.015370
Bard JA, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A (2018) Structure and function of the 26S proteasome. Annu Rev Biochem 87:697–724. https://doi.org/10.1146/annurev-biochem-062917-011931
Shin JY, Muniyappan S, Tran NN, Park H, Lee SB, Lee BH (2020) Deubiquitination reactions on the proteasome for proteasome versatility. Int J Mol Sci 21:5312. https://doi.org/10.3390/ijms21155312
Highet B, Dieriks BV, Murray HC, Faull RL, Curtis MA (2020) Huntingtin Aggregates in the Olfactory Bulb in Huntington’s Disease. Front Aging Neurosci 12:261. https://doi.org/10.3389/fnagi.2020.00261
Mogk A, Bukau B, Kampinga HH (2018) Cellular handling of protein aggregates by disaggregation machines. Mol Cell 69:214–226. https://doi.org/10.1016/j.molcel.2018.01.004
Makletsova MG, Syatkin SP, Poleshchuk VV, Urazgildeeva GR, Chigaleychik LA, Sungrapova CY, Illarioshkin SN (2019) Polyamines in Parkinson’s disease: their role in oxidative stress induction and protein aggregation. J Neurol Res 9:1–7. https://doi.org/10.14740/jnr509
Kouli A, Torsney KM, Kuan WL (2018) Parkinson’s disease: etiology, neuropathology, and pathogenesis. Exon Publications 21:3–26. https://doi.org/10.15586/codonpublications.parkinsonsdisease.2018.ch1
McCormack A, Keating DJ, Chegeni N, Colella A, Wang JJ, Chataway T (2019) Abundance of synaptic vesicle-related proteins in alpha-synuclein-containing protein inclusions suggests a targeted formation mechanism. Neurotox Res 35:883–897. https://doi.org/10.1007/s12640-019-00014-0
Matsumoto G, Inobe T, Amano T, Murai K, Nukina N, Mori N (2018) N-Acyldopamine induces aggresome formation without proteasome inhibition and enhances protein aggregation via p62/SQSTM1 expression. Sci Rep 8:1–4. https://doi.org/10.1038/s41598-018-27872-6
Meyer HJ, Rape M (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157:910–921. https://doi.org/10.1016/j.cell.2014.03.037
Varshavsky A (2017) The ubiquitin system, autophagy, and regulated protein degradation. Annu Rev Biochem 86:123–128. https://doi.org/10.1146/annurev-biochem-061516-044859
Hrelia P, Sita G, Ziche M, Ristori E, Marino A, Cordaro M, Molteni R, Spero V, Malaguti M, Morroni F, Hrelia S (2020) Common protective strategies in neurodegenerative disease: focusing on risk factors to target the cellular redox system. Oxidative Med Cell Longev 2020. https://doi.org/10.1155/2020/8363245
Upadhyay A, Sundaria N, Dhiman R, Prajapati VK, Prasad A, Mishra A (2021) Complex Inclusion Bodies and Defective Proteome Hubs in Neurodegenerative Disease: New Clues, New Challenges. Neuroscientist 3:1073858421989582. https://doi.org/10.1177/1073858421989582
Appel-Cresswell S, Vilarino-Guell C, Encarnacion M, Sherman H, Yu I, Shah B, Weir D, Thompson C, Szu-Tu C, Trinh J, Aasly JO (2013) Alpha-synuclein p. H50Q, a novel pathogenic mutation for Parkinson's disease. Mov Disord 28:811–813. https://doi.org/10.1002/mds.25421
Chen H, Huang X, Yuan L, Xia H, Xu H, Yang Y, Zheng W, Deng H (2016) A homozygous parkin p. G284R mutation in a Chinese family with autosomal recessive juvenile parkinsonism. Neurosci Lett 624:100–104. https://doi.org/10.1016/j.neulet.2016.05.011
Taipa R, Pereira C, Reis I, Alonso I, Bastos-Lima A, Melo-Pires M, Magalhaes M (2016) DJ-1 linked parkinsonism (PARK7) is associated with Lewy body pathology. Brain 139:1680–1687. https://doi.org/10.1093/brain/aww080
Bellomo G, Paciotti S, Gatticchi L, Parnetti L (2020) The vicious cycle between α-synuclein aggregation and autophagic-lysosomal dysfunction. Mov Disord 35:34–44. https://doi.org/10.1002/mds.27895
Ristic G, Tsou WL, Todi SV (2014) An optimal ubiquitin-proteasome pathway in the nervous system: the role of deubiquitinating enzymes. Front Mol Neurosci 7:72. https://doi.org/10.3389/fnmol.2014.00072
Wahl C, Kautzmann S, Krebiehl G, Strauss K, Woitalla D, Müller T, Bauer P, Riess O, Krüger R (2008) A comprehensive genetic study of the proteasomal subunit S6 ATPase in German Parkinson’s disease patients. J Neural Transm 115:1141. https://doi.org/10.1007/s00702-008-0054-3
Paine SM, Anderson G, Bedford K, Lawler K, Mayer RJ, Lowe J, Bedford L (2013) Pale body-like inclusion formation and neurodegeneration following depletion of 26S proteasomes in mouse brain neurones are independent of α-synuclein. PLoS One 8:e54711. https://doi.org/10.1371/journal.pone.0054711
Tofaris GK, Layfield R, Spillantini MG (2001) α-Synuclein metabolism and aggregation is linked to ubiquitin-independent degradation by the proteasome. FEBS Lett 509:22–26. https://doi.org/10.1016/S0014-5793(01)03115-5
Inamdar NN, Arulmozhi DK, Tandon A, Bodhankar SL (2007) Parkinson's disease: genetics and beyond. Curr Neuropharmacol 5:99–113. https://doi.org/10.2174/157015907780866893
Rao G, Croft B, Teng C, Awasthi V (2015) Ubiquitin-Proteasome System in Neurodegenerative Disorders. J Drug Metab Toxicol 2:1–6. https://doi.org/10.4172/2157-7609.1000187
Bentea E, Van der Perren A, Van Liefferinge J, El Arfani A, Albertini G, Demuyser T, Merckx E, Michotte Y, Smolders I, Baekelandt V, Massie A (2015) Nigral proteasome inhibition in mice leads to motor and non-motor deficits and increased expression of Ser129 phosphorylated α-synuclein. Front Behav Neurosci 9:68. https://doi.org/10.3389/fnbeh.2015.00068
de Vries RL, Przedborski S (2013) Mitophagy and Parkinson's disease: be eaten to stay healthy. Mol Cell Neurosci 55:37–43. https://doi.org/10.1016/j.mcn.2012.07.008
Klein C, Lohmann-Hedrich K (2007) Impact of recent genetic findings in Parkinson's disease. Curr Opin Neurol 20:453–464. https://doi.org/10.1097/WCO.0b013e3281e6692b
Leinartaité L, Svenningsson P (2017) Folding underlies bidirectional role of GPR37/Pael-R in Parkinson disease. Trends Pharmacol Sci 38:749–760. https://doi.org/10.1016/j.tips.2017.05.006
Tang MY, Vranas M, Krahn AI, Pundlik S, Trempe JF, Fon EA (2017) Structure-guided mutagenesis reveals a hierarchical mechanism of Parkin activation. Nat Commun 8:1–4. https://doi.org/10.1038/ncomms14697
Dove KK, Klevit RE, Rittinger K (2015) pUBLically unzipping Parkin: how phosphorylation exposes a ligase bit by bit. EMBO J 34:2486–2488. https://doi.org/10.15252/embj.201592857
Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, Dehejia A (1998) The ubiquitin pathway in Parkinson's disease. Nature 395:451–452. https://doi.org/10.1038/26652
Maraganore DM, Farrer MJ, Hardy JA, Lincoln SJ, McDonnell SK, Rocca WA (1999) Case-control study of the ubiquitin carboxy-terminal hydrolase L1 gene in Parkinson’s disease. Neurology 1:1858. https://doi.org/10.1212/WNL.53.8.1858
Sun F, Kanthasamy A, Anantharam V, Kanthasamy AG (2007) Environmental neurotoxic chemicals-induced ubiquitin proteasome system dysfunction in the pathogenesis and progression of Parkinson's disease. Pharmacol Ther 114:327–344. https://doi.org/10.1016/j.pharmthera.2007.04.001
Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J (2004) Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem 279:18614–18622. https://doi.org/10.1074/jbc.M401135200
Snyder NA, Silva GM (2021) Deubiquitinating enzymes (DUBs): Regulation, homeostasis, and oxidative stress response. J Biol Chem 297:101077. https://doi.org/10.1016/j.jbc.2021.101077
Matsuda N, Tanaka K (2010) Does impairment of the ubiquitin-proteasome system or the autophagy-lysosome pathway predispose individuals to neurodegenerative disorders such as Parkinson's disease? J Alzheimers Dis 19:1–9. https://doi.org/10.3233/JAD-2010-1231
Maraganore DM, Lesnick TG, Elbaz A, Chartier-Harlin MC, Gasser T, Krüger R, Hattori N, Mellick GD, Quattrone A, Satoh JI, Toda T (2004) UCHL1 is a Parkinson's disease susceptibility gene. Ann Neurol 55:512–521. https://doi.org/10.1002/ana.20017
Healy DG, Abou-Sleiman PM, Casas JP, Ahmadi KR, Lynch T, Gandhi S, Muqit MM, Foltynie T, Barker R, Bhatia KP, Quinn NP (2006) UCHL-1 is not a Parkinson's disease susceptibility gene. Ann Neurol 59:627–633. https://doi.org/10.1002/ana.20757
Bishop P, Rocca D, Henley JM (2016) Ubiquitin C-terminal hydrolase L1 (UCH-L1): structure, distribution and roles in brain function and dysfunction. Biochem J 473:2453–2462. https://doi.org/10.1042/BCJ20160082
Wang KK, Yang Z, Sarkis G, Torres I, Raghavan V (2017) Ubiquitin C-terminal hydrolase-L1 (UCH-L1) as a therapeutic and diagnostic target in neurodegeneration, neurotrauma and neuro-injuries. Expert Opin Ther Targets 21:627–638. https://doi.org/10.1080/14728222.2017.1321635
Andersson FI, Werrell EF, McMorran L, Crone WJ, Das C, Hsu ST, Jackson SE (2011) The effect of Parkinson's-disease-associated mutations on the deubiquitinating enzyme UCH-L1. J Mol Biol 407:261–272. https://doi.org/10.1016/j.jmb.2010.12.029
Bose A, Beal MF (2016) Mitochondrial dysfunction in Parkinson's disease. J Neurochem 139:216–231. https://doi.org/10.1111/jnc.13731
Ono K, Ikeda T, Takasaki JI, Yamada M (2011) Familial Parkinson disease mutations influence α-synuclein assembly. Neurobiol Dis 43:715–724. https://doi.org/10.1016/j.nbd.2011.05.025
Sun F, Anantharam V, Latchoumycandane C, Kanthasamy A, Kanthasamy AG (2005) Dieldrin induces ubiquitin-proteasome dysfunction in α-synuclein overexpressing dopaminergic neuronal cells and enhances susceptibility to apoptotic cell death. J Pharmacol Exp Ther 315:69–79. https://doi.org/10.1124/jpet.105.084632
Maries E, Dass B, Collier TJ, Kordower JH, Steece-Collier K (2003) The role of α-synuclein in Parkinson's disease: insights from animal models. Nat Rev Neurosci 4:727–738. https://doi.org/10.1038/nrn1199
Dehay B, Vila M, Bezard E, Brundin P, Kordower JH (2016) Alpha-synuclein propagation: new insights from animal models. Mov Disord 31:161–168. https://doi.org/10.1002/mds.26370
Cummins N, Tweedie A, Zuryn S, Bertran-Gonzalez J, Götz J (2019) Disease-associated tau impairs mitophagy by inhibiting Parkin translocation to mitochondria. EMBO J 38:e99360. https://doi.org/10.15252/embj.201899360
Perez FA, Palmiter RD (2005) Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci 102:2174–2179. https://doi.org/10.1073/pnas.0409598102
und Halbach OV, Schober A, Krieglstein K (2004) Genes, proteins, and neurotoxins involved in Parkinson’s disease. Prog Neurobiol 73:151-177. https://doi.org/10.1016/j.pneurobio.2004.05.002
Berko D, Herkon O, Braunstein I, Isakov E, David Y, Ziv T, Navon A, Stanhill A (2014) Inherent asymmetry in the 26S proteasome is defined by the ubiquitin receptor RPN13. J Biol Chem 289:5609–5618. https://doi.org/10.1074/jbc.M113.509380
Alexopoulou Z, Lang J, Perrett RM, Elschami M, Hurry ME, Kim HT, Mazaraki D, Szabo A, Kessler BM, Goldberg AL, Ansorge O (2016) Deubiquitinase Usp8 regulates α-synuclein clearance and modifies its toxicity in Lewy body disease. Proc Natl Acad Sci 113:E4688–E4697. https://doi.org/10.1073/pnas.1523597113
Zucchelli S, Codrich M, Marcuzzi F, Pinto M, Vilotti S, Biagioli M, Ferrer I, Gustincich S (2010) TRAF6 promotes atypical ubiquitination of mutant DJ-1 and alpha-synuclein and is localized to Lewy bodies in sporadic Parkinson's disease brains. Hum Mol Genet 19:3759–3770. https://doi.org/10.1093/hmg/ddq290
Wakabayashi K, Tanji K, Odagiri S, Miki Y, Mori F, Takahashi H (2013) The Lewy body in Parkinson’s disease and related neurodegenerative disorders. Mol Neurobiol 47:495–508. https://doi.org/10.1007/s12035-012-8280-y
Franco-Iborra S, Vila M, Perier C (2018) Mitochondrial quality control in neurodegenerative diseases: focus on Parkinson's disease and Huntington's disease. Front Neurosci 12:342. https://doi.org/10.3389/fnins.2018.00342
Shin WH, Park JH, Chung KC (2020) The central regulator p62 between ubiquitin proteasome system and autophagy and its role in the mitophagy and Parkinson’s disease. BMB Rep 53:56. https://doi.org/10.5483/BMBRep.2020.53.1.283
Sugatani J, Noguchi Y, Hattori Y, Yamaguchi M, Yamazaki Y, Ikari A (2016) Threonine-408 regulates the stability of human pregnane X receptor through its phosphorylation and the CHIP/chaperone-autophagy pathway. Drug Metab Dispos 44:137–150. https://doi.org/10.1124/dmd.115.066308
El-Saiy KA, Sayed RH, El-Sahar AE, Kandil EA (2022) Modulation of histone deacetylase, the ubiquitin proteasome system, and autophagy underlies the neuroprotective effects of venlafaxine in a rotenone-induced Parkinson's disease model in rats. Chem Biol Interact 354:109841. https://doi.org/10.1016/j.cbi.2022.109841
Wang XL, Feng ST, Wang YT, Yuan YH, Li ZP, Chen NH, Wang ZZ, Zhang Y (2021) Mitophagy, a Form of Selective Autophagy, Plays an Essential Role in Mitochondrial Dynamics of Parkinson’s Disease. Cell Mol Neurobiol 2021:1–9. https://doi.org/10.1007/s10571-021-01039-w
Khaminets A, Behl C, Dikic I (2016) Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol 26(1):6–16. https://doi.org/10.1016/j.tcb.2015.08.010
Huang CY, Sivalingam K, Shibu MA, Liao PH, Ho TJ, Kuo WW, Chen RJ, Day CH, Viswanadha VP, Ju DT (2020) Induction of autophagy by vasicinone protects neural cells from mitochondrial dysfunction and attenuates paraquat-mediated Parkinson’s disease associated α-synuclein levels. Nutrients 12:1707. https://doi.org/10.3390/nu12061707
Shen YF, Tang Y, Zhang XJ, HuANG KX, Le WD (2013) Adaptive changes in autophagy after UPS impairment in Parkinson's disease. Acta Pharmacol Sin 34(5):667–673. https://doi.org/10.1038/aps.2012.203
Rango M, Bresolin N (2018) Brain mitochondria, aging, and Parkinson’s disease. Genes 9:250. https://doi.org/10.3390/genes9050250
Cheng J, Deng Y, Zhou J (2021) Role of the Ubiquitin System in Chronic Pain. Front Mol Neurosci 14:674914. https://doi.org/10.3389/fnmol.2021.674914
Ross CA, Pickart CM (2004) The ubiquitin–proteasome pathway in Parkinson's disease and other neurodegenerative diseases. Trends Cell Biol 14:703–711. https://doi.org/10.1016/j.tcb.2004.10.006
Gupta N (2014) Ubiquitin-proteasome system modulates platelet function, Cleveland State University https://engagedscholarship.csuohio.edu/etdarchive/115
Olanow CW, McNaught KS (2006) Ubiquitin–proteasome system and Parkinson's disease. Mov Disord 21:1806–1823. https://doi.org/10.1002/mds.21013
Moskal N, Riccio V, Bashkurov M, Taddese R, Datti A, Lewis PN, McQuibban GA (2020) ROCK inhibitors upregulate the neuroprotective Parkin-mediated mitophagy pathway. Nat Commun 11:1–4. https://doi.org/10.1038/s41467-019-13781-3
Ge P, Dawson VL, Dawson TM (2020) PINK1 and Parkin mitochondrial quality control: A source of regional vulnerability in Parkinson’s disease. Mol Neurodegener 15:1–8. https://doi.org/10.1186/s13024-020-00367-7
Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW (2003) Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol 23:8786–8794. https://doi.org/10.1128/MCB.23.23.8786-8794.2003
Crider A, Pandya CD, Peter D, Ahmed AO, Pillai A (2014) Ubiquitin-proteasome dependent degradation of GABA A α1 in autism spectrum disorder. Mol Autism 5:1-0. https://doi.org/10.1186/2040-2392-5-45
Bareggi SR, Cornelli U (2012) Clioquinol: review of its mechanisms of action and clinical uses in neurodegenerative disorders. CNS Neurosci Ther 18:41–46. https://doi.org/10.1111/j.1755-5949.2010.00231.x
Stefanova N, Kaufmann WA, Humpel C, Poewe W, Wenning GK (2012) Systemic proteasome inhibition triggers neurodegeneration in a transgenic mouse model expressing human α-synuclein under oligodendrocyte promoter: implications for multiple system atrophy. Acta Neuropathol 124:51–65. https://doi.org/10.1007/s00401-012-0977-5
Ma Y, Xu B, Fang Y, Yang Z, Cui J, Zhang L, Zhang L (2011) Synthesis and SAR study of novel peptide aldehydes as inhibitors of 20S proteasome. Molecules 16:7551–7564. https://doi.org/10.3390/molecules16097551
Bir A, Sen O, Anand S, Khemka VK, Banerjee P, Cappai R, Sahoo A, Chakrabarti S (2014) α-synuclein-induced mitochondrial dysfunction in isolated preparation and intact cells: Implications in the pathogenesis of Parkinson's disease. J Neurochem 131:868–877. https://doi.org/10.1111/jnc.12966
Zhou Z, Kerk S, Meng Lim T (2008) Endogenous dopamine (DA) renders dopaminergic cells vulnerable to challenge of proteasome inhibitor MG132. Free Radic Res 42:456–466. https://doi.org/10.1080/10715760802005177
Jung EB, Lee CS (2014) Baicalein attenuates proteasome inhibition-induced apoptosis by suppressing the activation of the mitochondrial pathway and the caspase-8-and Bid-dependent pathways. Eur J Pharmacol 730:116–124. https://doi.org/10.1016/j.ejphar.2014.02.039
Hegde AN, Haynes KA, Bach SV, Beckelman BC (2014) Local ubiquitin-proteasome-mediated proteolysis and long-term synaptic plasticity. Front Mol Neurosci 7:96. https://doi.org/10.3389/fnmol.2014.00096
Snider BJ, Tee LY, Canzoniero LM, Babcock DJ, Choi DW (2002) NMDA antagonists exacerbate neuronal death caused by proteasome inhibition in cultured cortical and striatal neurons. Eur J Neurosci 15:419–428. https://doi.org/10.1046/j.0953-816x.2001.01867.x
Höglinger GU, Carrard G, Michel PP, Medja F, Lombès A, Ruberg M, Friguet B, Hirsch EC (2003) Dysfunction of mitochondrial complex I and the proteasome: interactions between two biochemical deficits in a cellular model of Parkinson's disease. J Neurochem 86:1297–1307. https://doi.org/10.1046/j.1471-4159.2003.01952.x
Chang KH, Lee-Chen GJ, Wu YR, Chen YJ, Lin JL, Li M, Chen IC, Lo YS, Wu HC, Chen CM (2016) Impairment of proteasome and anti-oxidative pathways in the induced pluripotent stem cell model for sporadic Parkinson's disease. Parkinsonism Relat Disord 24:81–88. https://doi.org/10.1016/j.parkreldis.2016.01.001
Filatova EV, Shadrina MI, Alieva AK, Kolacheva AA, Slominsky PA, Ugrumov MV (2014) Expression analysis of genes of ubiquitin-proteasome protein degradation system in MPTP-induced mice models of early stages of Parkinson’s disease. Dokl Biochem Biophys 116-118. https://doi.org/10.1134/S1607672914030107
Whitworth AJ, Theodore DA, Greene JC, Beneš H, Wes PD, Pallanck LJ (2005) Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson's disease. Proc Natl Acad Sci 102:8024–8029. https://doi.org/10.1073/pnas.0501078102
Madsen DA, Schmidt SI, Blaabjerg M, Meyer M (2021) Interaction between Parkin and α-Synuclein in PARK2-Mediated Parkinson’s Disease. Cells 10:283. https://doi.org/10.3390/cells10020283
Fabbri M, Perez-Lloret S, Rascol O (2020) Therapeutic strategies for Parkinson’s disease: promising agents in early clinical development. Expert Opin Investig Drugs 29:1249–1267. https://doi.org/10.1080/13543784.2020.1814252
Akhtar J, Wang Z, Zhang ZP, Bi MM (2013) Lentiviral-mediated RNA interference targeting stathmin1 gene in human gastric cancer cells inhibits proliferation in vitro and tumor growth in vivo. J Transl Med 11:1–9. https://doi.org/10.1186/1479-5876-11-212
Shen HY, He JC, Wang Y, Huang QY, Chen JF (2005) Geldanamycin induces heat shock protein 70 and protects against MPTP-induced dopaminergic neurotoxicity in mice. J Biol Chem 280:39962–39969. https://doi.org/10.1074/jbc.M505524200
Opattova A, Cente M, Novak M, Filipcik P (2015) The ubiquitin proteasome system as a potential therapeutic target for treatment of neurodegenerative diseases. Gen Physiol Biophys 34:337–352. https://doi.org/10.4149/gpb_2015024
Dantuma NP, Bott LC (2014) The ubiquitin-proteasome system in neurodegenerative diseases: precipitating factor, yet part of the solution. Front Mol Neurosci 7:70. https://doi.org/10.3389/fnmol.2014.00070
Jeon J, Kim W, Jang J, Isacson O, Seo H (2016) Gene therapy by proteasome activator, PA28γ, improves motor coordination and proteasome function in Huntington’s disease YAC128 mice. Neuroscience 324:20–28. https://doi.org/10.1016/j.neuroscience.2016.02.054
Jung T, Grune T (2013) The proteasome and the degradation of oxidized proteins: part I—structure of proteasomes. Redox boil 1:178–182. https://doi.org/10.1016/j.redox.2013.01.004
Lehrbach NJ, Breen PC, Ruvkun G (2019) Protein sequence editing of SKN-1A/Nrf1 by peptide: N-glycanase controls proteasome gene expression. Cell 177:737–750. https://doi.org/10.1016/j.cell.2019.03.035
Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues AP, Manning G, Dillin A (2012) RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489:263–268. https://doi.org/10.1038/nature11315
Raina K, Crews CM (2017) Targeted protein knockdown using small molecule degraders. Curr Opin Chem Biol 39:46–53. https://doi.org/10.1016/j.cbpa.2017.05.016
Schneekloth AR, Pucheault M, Tae HS, Crews CM (2008) Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg Med Chem Lett 18:5904–5908. https://doi.org/10.1016/j.bmcl.2008.07.114
Bondeson DP, Smith BE, Burslem GM, Buhimschi AD, Hines J, Jaime-Figueroa S, Wang J, Hamman BD, Ishchenko A, Crews CM (2018) Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem Biol 25:78–87. https://doi.org/10.1016/j.chembiol.2017.09.010
Lucas X, Ciulli A (2017) Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies. Curr Opin Struct Biol 44:101–110. https://doi.org/10.1016/j.sbi.2016.12.015
Au YZ, Wang T, Sigua LH, Qi J (2020) Peptide-based PROTAC: the predator of pathological proteins. Cell Chem Biol 27:637–639. https://doi.org/10.1016/j.chembiol.2020.06.002
Konstantinidou M, Oun A, Pathak P, Zhang B, Wang Z, Ter Brake F, Dolga AM, Kortholt A, Dömling A (2021) The tale of proteolysis targeting chimeras (PROTACs) for Leucine-Rich Repeat Kinase 2 (LRRK2). ChemMedChem 16:959. https://doi.org/10.1002/cmdc.202000872
Silva MC, Ferguson FM, Cai Q, Donovan KA, Nandi G, Patnaik D, Zhang T, Huang HT, Lucente DE, Dickerson BC, Mitchison TJ (2019) Targeted degradation of aberrant tau in frontotemporal dementia patient-derived neuronal cell models. Elife 8:e45457. https://doi.org/10.7554/eLife.45457
Smith MH, Ploegh HL, Weissman JS (2011) Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334:1086–1090. https://doi.org/10.1126/science.1209235
Ohtake F, Baba A, Takada I, Okada M, Iwasaki K, Miki H, Takahashi S, Kouzmenko A, Nohara K, Chiba T, Fujii-Kuriyama Y (2007) Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature 446:562–566. https://doi.org/10.1038/nature05683
Kawajiri K, Kobayashi Y, Ohtake F, Ikuta T, Matsushima Y, Mimura J, Pettersson S, Pollenz RS, Sakaki T, Hirokawa T, Akiyama T (2009) Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc Natl Acad Sci 106:13481–13486. https://doi.org/10.1073/pnas.0902132106
Luecke-Johansson S, Gralla M, Rundqvist H, Ho JC, Johnson RS, Gradin K, Poellinger L (2017) A molecular mechanism to switch the aryl hydrocarbon receptor from a transcription factor to an E3 ubiquitin ligase. Mol Cell Biol 37:e00630–e00616. https://doi.org/10.1128/MCB.00630-16
Rana R, Coulter S, Kinyamu H, Goldstein JA (2013) RBCK1, an E3 ubiquitin ligase, interacts with and ubiquinates the human pregnane X receptor. Drug Metab Dispos 41:398–405. https://doi.org/10.1124/dmd.112.048728
Acknowledgment
The authors would like to thank Chitkara College of Pharmacy, Chitkara University, Punjab, India, for providing the various resources for completion of the current article.
Funding
The current article did not receive any funding.
Author information
Authors and Affiliations
Contributions
TB and SK conceived the idea and wrote the first draft; ZMA, SS, NS and VNB reviewed the literature; AS and SY worked on figures; SB, AAH, and YA edited the manuscript; TB, MAA, and SBU did proof reading.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• In the aetiology of Parkinson's disease, the vicious cycle between proteasome failure and protein accumulation is crucial.
• The ubiquitin–proteasome system (UPS) is the primary hydrolytic system involved in the bulk of protein breakdown.
• PROTAC's development brings up new possibilities for treating Parkinson's disease through proteasome modulation.
• Proteasome homoeostasis regulation is a promising treatment for Parkinson's disease.
Rights and permissions
About this article
Cite this article
Behl, T., Kumar, S., Althafar, Z.M. et al. Exploring the Role of Ubiquitin–Proteasome System in Parkinson's Disease. Mol Neurobiol 59, 4257–4273 (2022). https://doi.org/10.1007/s12035-022-02851-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-02851-1