Abstract
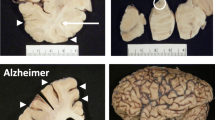
Increased amyloid β (Aβ) aggregation is a hallmark feature of Alzheimer’s disease (AD) pathology. The APP/PS1 mouse model of AD exhibits accumulation of Aβ in the retina and demonstrates reduced retinal function and other degenerative changes. The overall molecular effects of AD pathology on the retina remain undetermined. Using a proteomics approach, this study assessed the molecular effects of Aβ accumulation and progression of AD pathology on the retina. Retinal tissues from younger (2.5 months) and older 8-month APP/PS1 mice were analysed for protein expression changes. A multiplexed proteomics approach using chemical isobaric tandem mass tags was applied followed by functional and protein-protein interaction analyses using Ingenuity pathway (IPA) and STRING computational tools. We identified approximately 2000 proteins each in the younger (upregulated 50; downregulated 36) and older set of APP/PS1 (upregulated 85; downregulated 79) mice retinas. Amyloid precursor protein (APP) was consistently upregulated two to threefold in both younger and older retinas (p < 0.0001). Mass spectrometry data further revealed that older APP/PS1 mice retinas had elevated levels of proteolytic enzymes cathepsin D, presenilin 2 and nicastrin that are associated with APP processing. Increased levels of proteasomal proteins Psma5, Psmd3 and Psmb2 were also observed in the older AD retinas. In contrast to the younger animals, significant downregulation of protein synthesis and elongation associated proteins such as Eef1a1, Rpl35a, Mrpl2 and Eef1e1 (p < 0.04) was identified in the older mice retinas. This study reports for the first time that not only old but also young APP/PS1 animals demonstrate increased amyloid protein levels in their retinas. Quantitative proteomics reveals new molecular insights which may represent a cellular response to clear amyloid build-up. Further, downregulation of ribosomal proteins involved in protein biosynthesis was observed which might be considered a toxicity effect.









Similar content being viewed by others
References
Group, G. B. D. N. D. C (2017) Global, regional, and national burden of neurological disorders during 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Neurol 16:877–897. https://doi.org/10.1016/S1474-4422(17)30299-5
Hebert LE, Weuve J, Scherr PA, Evans DA (2013) Alzheimer disease in the United States (2010-2050) estimated using the 2010 census. Neurology 80:1778–1783. https://doi.org/10.1212/WNL.0b013e31828726f5
Bilgel M et al (2016) Individual estimates of age at detectable amyloid onset for risk factor assessment. Alzheimers Dement 12:373–379. https://doi.org/10.1016/j.jalz.2015.08.166
Villemagne VL et al (2013) Amyloid beta deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer's disease: a prospective cohort study. Lancet Neurol 12:357–367. https://doi.org/10.1016/S1474-4422(13)70044-9
Chi S, Yu JT, Tan MS, Tan L (2014) Depression in Alzheimer's disease: epidemiology, mechanisms, and management. J Alzheimers Dis 42:739–755. https://doi.org/10.3233/JAD-140324
Wu N, Rao X, Gao Y, Wang J, Xu F (2013) Amyloid-beta deposition and olfactory dysfunction in an Alzheimer's disease model. J Alzheimers Dis 37:699–712. https://doi.org/10.3233/JAD-122443
Gupta VK et al (2016) Amyloid beta accumulation and inner retinal degenerative changes in Alzheimer's disease transgenic mouse. Neurosci Lett 623:52–56. https://doi.org/10.1016/j.neulet.2016.04.059
Holth J, Patel T, Holtzman DM (2017) Sleep in Alzheimer's disease - beyond amyloid. Neurobiol Sleep Circadian Rhythms 2:4–14. https://doi.org/10.1016/j.nbscr.2016.08.002
Golzan SM et al (2017) Retinal vascular and structural changes are associated with amyloid burden in the elderly: ophthalmic biomarkers of preclinical Alzheimer's disease. Alzheimers Res Ther 9:13. https://doi.org/10.1186/s13195-017-0239-9
Criscuolo C et al (2018) The retina as a window to early dysfunctions of Alzheimer's disease following studies with a 5xFAD mouse model. Neurobiol Aging 67:181–188. https://doi.org/10.1016/j.neurobiolaging.2018.03.017
Perez SE, Lumayag S, Kovacs B, Mufson EJ, Xu S (2009) Beta-amyloid deposition and functional impairment in the retina of the APPswe/PS1DeltaE9 transgenic mouse model of Alzheimer's disease. Invest Ophthalmol Vis Sci 50:793–800. https://doi.org/10.1167/iovs.08-2384
Liu B et al (2009) Amyloid-peptide vaccinations reduce {beta}-amyloid plaques but exacerbate vascular deposition and inflammation in the retina of Alzheimer's transgenic mice. Am J Pathol 175:2099–2110. https://doi.org/10.2353/ajpath.2009.090159
Berisha F, Feke GT, Trempe CL, McMeel JW, Schepens CL (2007) Retinal abnormalities in early Alzheimer's disease. Invest Ophthalmol Vis Sci 48:2285–2289. https://doi.org/10.1167/iovs.06-1029
Sivak JM (2013) The aging eye: common degenerative mechanisms between the Alzheimer's brain and retinal disease. Invest Ophthalmol Vis Sci 54:871–880. https://doi.org/10.1167/iovs.12-10827
M Mirzaei VBG, Gupta VK (2018) Retinal changes in Alzheimer's disease: disease mechanisms to evaluation perspectives. J Neurol Neuromed 3:11–13
Sperling RA et al (2011) Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 7:280–292. https://doi.org/10.1016/j.jalz.2011.03.003
Buchhave P et al (2012) Cerebrospinal fluid levels of beta-amyloid 1-42, but not of tau, are fully changed already 5 to 10 years before the onset of Alzheimer dementia. Arch Gen Psychiatry 69:98–106. https://doi.org/10.1001/archgenpsychiatry.2011.155
Johnson KA et al (2013) Appropriate use criteria for amyloid PET: a report of the Amyloid Imaging Task Force, the Society of Nuclear Medicine and Molecular Imaging, and the Alzheimer's Association. Alzheimers Dement 9:e-1–e16. https://doi.org/10.1016/j.jalz.2013.01.002
Hart NJ, Koronyo Y, Black KL, Koronyo-Hamaoui M (2016) Ocular indicators of Alzheimer's: exploring disease in the retina. Acta Neuropathol 132:767–787. https://doi.org/10.1007/s00401-016-1613-6
Chiu K et al (2012) Neurodegeneration of the retina in mouse models of Alzheimer's disease: what can we learn from the retina? Age (Dordr) 34:633–649. https://doi.org/10.1007/s11357-011-9260-2
Gupta V et al (2016) One protein, multiple pathologies: multifaceted involvement of amyloid beta in neurodegenerative disorders of the brain and retina. Cell Mol Life Sci 73:4279–4297. https://doi.org/10.1007/s00018-016-2295-x
Gasparini L et al (2011) Tau inclusions in retinal ganglion cells of human P301S tau transgenic mice: effects on axonal viability. Neurobiol Aging 32:419–433. https://doi.org/10.1016/j.neurobiolaging.2009.03.002
Ning A, Cui J, To E, Ashe KH, Matsubara J (2008) Amyloid-beta deposits lead to retinal degeneration in a mouse model of Alzheimer disease. Invest Ophthalmol Vis Sci 49:5136–5143. https://doi.org/10.1167/iovs.08-1849
Mirzaei M et al (2017) Age-related neurodegenerative disease associated pathways identified in retinal and vitreous proteome from human glaucoma eyes. Sci Rep 7:12685. https://doi.org/10.1038/s41598-017-12858-7
Serneels L et al (2009) Gamma-Secretase heterogeneity in the Aph1 subunit: relevance for Alzheimer's disease. Science 324:639–642. https://doi.org/10.1126/science.1171176
Gengler S, Hamilton A, Holscher C (2010) Synaptic plasticity in the hippocampus of a APP/PS1 mouse model of Alzheimer's disease is impaired in old but not young mice. PLoS One 5:e9764. https://doi.org/10.1371/journal.pone.0009764
Radde R et al (2006) Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep 7:940–946. https://doi.org/10.1038/sj.embor.7400784
Rupp NJ, Wegenast-Braun BM, Radde R, Calhoun ME, Jucker M (2011) Early onset amyloid lesions lead to severe neuritic abnormalities and local, but not global neuron loss in APPPS1 transgenic mice. Neurobiol Aging 32(2324):e2321–e2326. https://doi.org/10.1016/j.neurobiolaging.2010.08.014
Winkler BS (1972) The electroretinogram of the isolated rat retina. Vision Res 12:1183–1198
Gupta V et al (2017) Glaucoma is associated with plasmin proteolytic activation mediated through oxidative inactivation of neuroserpin. Sci Rep 7:8412. https://doi.org/10.1038/s41598-017-08688-2
Mirzaei M et al (2017) in Proteome bioinformatics (eds Shivakumar Keerthikumar & Suresh Mathivanan) 45-66 (Springer New York)
Margolin AA et al (2009) Empirical Bayes analysis of quantitative proteomics experiments. PloS one 4:e7454. https://doi.org/10.1371/journal.pone.0007454
Kammers K, Cole RN, Tiengwe C, Ruczinski I (2015) Detecting significant changes in protein abundance. EuPA open Proteomics 7:11–19. https://doi.org/10.1016/j.euprot.2015.02.002
Gupta VK, Rajala A, Daly RJ, Rajala RV (2010) Growth factor receptor-bound protein 14: a new modulator of photoreceptor-specific cyclic-nucleotide-gated channel. EMBO Rep 11:861–867. https://doi.org/10.1038/embor.2010.142
Gupta VK, Rajala A, Rajala RV (2012) Insulin receptor regulates photoreceptor CNG channel activity. Am J Physiol Endocrinol Metab 303:E1363–E1372. https://doi.org/10.1152/ajpendo.00199.2012
Chitranshi N et al (2017) Ptpn11 induces endoplasmic stress and apoptosis in Sh-Sy5y cells. Neuroscience 364:175–189. https://doi.org/10.1016/j.neuroscience.2017.09.028
Rajala A, Gupta VK, Anderson RE, Rajala RV (2013) Light activation of the insulin receptor regulates mitochondrial hexokinase. A possible mechanism of retinal neuroprotection. Mitochondrion 13:566–576. https://doi.org/10.1016/j.mito.2013.08.005
Chitranshi N et al (2018) Loss of Shp2 rescues BDNF/TrkB signaling and contributes to improved retinal ganglion cell neuroprotection. Mol Ther. https://doi.org/10.1016/j.ymthe.2018.09.019
Sultana R et al (2006) Identification of nitrated proteins in Alzheimer's disease brain using a redox proteomics approach. Neurobiol Dis 22:76–87. https://doi.org/10.1016/j.nbd.2005.10.004
Yu L et al (2018) Targeted brain proteomics uncover multiple pathways to Alzheimer's dementia. Ann Neurol 84:78–88. https://doi.org/10.1002/ana.25266
Kempf SJ et al (2016) An integrated proteomics approach shows synaptic plasticity changes in an APP/PS1 Alzheimer's mouse model. Oncotarget 7:33627–33648. https://doi.org/10.18632/oncotarget.9092
Koronyo Y et al (2017) Retinal amyloid pathology and proof-of-concept imaging trial in Alzheimer's disease. JCI Insight 2. https://doi.org/10.1172/jci.insight.93621
Johnson-Wood K et al (1997) Amyloid precursor protein processing and A beta42 deposition in a transgenic mouse model of Alzheimer disease. Proc Natl Acad Sci U S A 94:1550–1555
Oliveira JM, Henriques AG, Martins F, Rebelo S, da Cruz e Silva OA (2015) Amyloid-beta modulates both AbetaPP and Tau phosphorylation. J Alzheimers Dis 45:495–507. https://doi.org/10.3233/JAD-142664
Chiasseu M et al (2016) Tau Accumulation, altered phosphorylation, and missorting promote neurodegeneration in glaucoma. J Neurosci 36:5785–5798. https://doi.org/10.1523/JNEUROSCI.3986-15.2016
Zhao H et al (2013) Hyperphosphorylation of tau protein by calpain regulation in retina of Alzheimer's disease transgenic mouse. Neurosci Lett 551:12–16. https://doi.org/10.1016/j.neulet.2013.06.026
Pedros I et al (2014) Early alterations in energy metabolism in the hippocampus of APPswe/PS1dE9 mouse model of Alzheimer's disease. Biochim Biophys Acta 1842:1556–1566. https://doi.org/10.1016/j.bbadis.2014.05.025
Bennett RE et al (2017) Enhanced Tau aggregation in the presence of amyloid beta. Am J Pathol 187:1601–1612. https://doi.org/10.1016/j.ajpath.2017.03.011
Kranenburg O et al (2005) Amyloid-beta-stimulated plasminogen activation by tissue-type plasminogen activator results in processing of neuroendocrine factors. Neuroscience 131:877–886. https://doi.org/10.1016/j.neuroscience.2004.11.044
Tucker HM et al (2000) The plasmin system is induced by and degrades amyloid-beta aggregates. J Neurosci 20:3937–3946
Hook V et al (2005) Inhibition of cathepsin B reduces beta-amyloid production in regulated secretory vesicles of neuronal chromaffin cells: evidence for cathepsin B as a candidate beta-secretase of Alzheimer's disease. Biol Chem 386:931–940. https://doi.org/10.1515/BC.2005.108
Hook VY, Kindy M, Hook G (2008) Inhibitors of cathepsin B improve memory and reduce beta-amyloid in transgenic Alzheimer disease mice expressing the wild-type, but not the Swedish mutant, beta-secretase site of the amyloid precursor protein. J Biol Chem 283:7745–7753. https://doi.org/10.1074/jbc.M708362200
Embury CM et al (2017) Cathepsin B improves ss-amyloidosis and learning and memory in models of Alzheimer's disease. J Neuroimmune Pharmacol 12:340–352. https://doi.org/10.1007/s11481-016-9721-6
Pamren A et al (2011) Mutations in nicastrin protein differentially affect amyloid beta-peptide production and Notch protein processing. J Biol Chem 286:31153–31158. https://doi.org/10.1074/jbc.C111.235267
Sesele K et al (2013) Conditional inactivation of nicastrin restricts amyloid deposition in an Alzheimer's disease mouse model. Aging Cell 12:1032–1040. https://doi.org/10.1111/acel.12131
Nadler Y et al (2008) Increased expression of the gamma-secretase components presenilin-1 and nicastrin in activated astrocytes and microglia following traumatic brain injury. Glia 56:552–567. https://doi.org/10.1002/glia.20638
Ciechanover A, Kwon YT (2015) Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med 47:e147. https://doi.org/10.1038/emm.2014.117
Keck S, Nitsch R, Grune T, Ullrich O (2003) Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer's disease. J Neurochem 85:115–122
Morozov AV et al (2016) Amyloid-beta increases activity of proteasomes capped with 19S and 11S regulators. J Alzheimers Dis 54:763–776. https://doi.org/10.3233/JAD-160491
Aso E et al (2012) Amyloid generation and dysfunctional immunoproteasome activation with disease progression in animal model of familial Alzheimer's disease. Brain Pathol 22:636–653. https://doi.org/10.1111/j.1750-3639.2011.00560.x
Ding Q, Markesbery WR, Chen Q, Li F, Keller JN (2005) Ribosome dysfunction is an early event in Alzheimer's disease. J Neurosci Off J Soc Neurosci 25:9171–9175. https://doi.org/10.1523/JNEUROSCI.3040-05.2005
Hernandez-Ortega K, Garcia-Esparcia P, Gil L, Lucas JJ, Ferrer I (2016) Altered machinery of protein synthesis in Alzheimer's: from the nucleolus to the ribosome. Brain Pathol 26:593–605. https://doi.org/10.1111/bpa.12335
Garcia-Esparcia P et al (2017) Altered mechanisms of protein synthesis in frontal cortex in Alzheimer disease and a mouse model. Am J Neurodegener Dis 6:15–25
Meier S et al (2016) Pathological Tau promotes neuronal damage by impairing ribosomal function and decreasing protein synthesis. J Neurosci Off J Soc Neurosci 36:1001–1007. https://doi.org/10.1523/JNEUROSCI.3029-15.2016
Bou Samra E et al (2017) A role for Tau protein in maintaining ribosomal DNA stability and cytidine deaminase-deficient cell survival. Nat Commun 8:693. https://doi.org/10.1038/s41467-017-00633-1
Wu Y et al (2016) Regulation of global gene expression and cell proliferation by APP. Sci Rep 6:22460. https://doi.org/10.1038/srep22460
Mao JJ et al (2001) The relationship between alphaB-crystallin and neurofibrillary tangles in Alzheimer's disease. Neuropathol Appl Neurobiol 27:180–188
Dulle JE, Fort PE (2016) Crystallins and neuroinflammation: the glial side of the story. Biochim Biophys Acta 1860:278–286. https://doi.org/10.1016/j.bbagen.2015.05.023
Piri N, Kwong JM, Caprioli J (2013) Crystallins in retinal ganglion cell survival and regeneration. Mol Neurobiol 48:819–828. https://doi.org/10.1007/s12035-013-8470-2
Liedtke T, Schwamborn JC, Schroer U, Thanos S (2007) Elongation of axons during regeneration involves retinal crystallin beta b2 (crybb2). Mol Cell Proteomics 6:895–907. https://doi.org/10.1074/mcp.M600245-MCP200
Anders F et al (2017) Intravitreal injection of beta-crystallin B2 improves retinal ganglion cell survival in an experimental animal model of glaucoma. PloS one 12:e0175451. https://doi.org/10.1371/journal.pone.0175451
Shammas SL et al (2011) Binding of the molecular chaperone alphaB-crystallin to Abeta amyloid fibrils inhibits fibril elongation. Biophys J 101:1681–1689. https://doi.org/10.1016/j.bpj.2011.07.056
Bjorkdahl C et al (2008) Small heat shock proteins Hsp27 or alphaB-crystallin and the protein components of neurofibrillary tangles: tau and neurofilaments. J Neurosci Res 86:1343–1352. https://doi.org/10.1002/jnr.21589
Acknowledgements
We acknowledge the support from the Ophthalmic Research Institute of Australia, National Health and Medical Research Council (NHMRC) and Hillcrest Foundation. The mass spectrometry analysis in this study was conducted at the Australian Proteome Analysis Facility supported by the Australian Government’s National Collaborative Research Infrastructure Scheme (NCRIS).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Supplementary Figure 1:
Protein interaction and functional analysis. A, B. both young and older APP/PS1 mice respectively used to understand biochemical and physiological retinal anomalies cause due to AD progression with age. (PNG 395 kb)
Supplementary Figure 2:
Immunostaining of the old retinal sections from WT and APP/PS1 mice illustrating nicastrin (Red–Cy3) expression (blue-DAPI) (n=3). Overexpressed locations were indicated by the white arrowheads. Scale=50 μm. (PNG 261 kb)
Supplementary Figure 3:
Relative fluorescence intensity of the GCL region of immunostained sections as labelled above each graph. The sections were compared between WT and APP retinal staining. Aβ (Fig. 5C); Presenilin 1, Tau, pTau Ser404 (Fig. 6 B,C,D), Cathepsin B (Fig. 7B), PSMB2 (Fig. 7D), MRPL2 (Fig. 8B), EEF1E1 (Fig. 8D), αB Crystallin (Fig. 9B), βB2 Crystallin (Fig. 9D) and βB3 Crystallin (Fig. 9F), (n=3 each) (****p<0.0001, ***p<0.001, *p<0.01). (PNG 303 kb)
Supplementary Table 1:
The list of proteins identified in the young retinal samples, including differentially expressed proteins identified from pairwise comparison test of APP/Ps1 vs. WT. The table 1 contains 4 independent sheets; Sheet 1- list of quantified proteins in the TMT experiments number 1 (younger retina), Sheet 2- list of differentially expressed proteins (APP vs WT comparison student t-test, p-value ≤ 0.05 and ≥ 1.15-fold difference), Sheet 3 and Sheet 4- list of down-regulated protein and list of up-regulated proteins respectively. Column A represents: UniProt ID; Column B: Peptide counts; Column C-G: fold changes ratios obtained from the TMT reporter ion intensities of APP replicates (129N, 129C, 130 N, 130 C and 131) versus the reference replicate 126 (one of the WT replicates); Column H-K: fold change ratios obtained from the TMT reporter ion intensities of WT replicates (127N, 127C, 128N, 128C) versus the reference replicate 126 (one of the WT replicates); Column L: p-values generated from APP vs WT t-test comparison test (p-value ≤ 0.05 highlighted as yellow); Column M: Max fold change of mean ration of APP vs WT; Column N: mean value of the fold change ratio of APP replicates vs reference replicate (126), purple: up-regulated, green: down-regulated; Column O: mean value of the fold change ratio of WT replicates vs reference replicate (126), purple: up-regulated, green: down-regulated ; Column P: Protein name/description and Column Q: Gene ontology information of every protein. (XLSX 840 kb)
Supplementary Table 2:
The list of proteins identified in the old retina samples, including differentially expressed proteins from pairwise comparison test of APP/Ps1 vs. WT. The table 2 contains 4 independent sheets; Sheet 1-list of quantified proteins in the TMT experiments number 1 (older retina), Sheet 2- list of differentially expressed proteins (APP vs WT comparison student t-test, p-value ≤ 0.05 and ≥ 1.15-fold difference), Sheet 3 and 4- list of down-regulated protein and list of up-regulated proteins respectively. Column A represents : UniProt ID; Column B: Peptide counts; Column C-G: fold changes ratios obtained from the TMT reporter ion intensities of APP replicates (129N, 129C, 130 N, 130 C and 131) versus the reference replicate 126 (one of the WT replicates); Column H-K: fold change ratios obtained from the TMT reporter ion intensities of WT replicates (127N, 127C, 128N, 128C) versus the reference replicate 126 (one of the WT replicates); Column L: P-values generated from APP vs WT t-test comparison test (p-value ≤ 0.05 highlighted as yellow); Column M: Max fold change of mean ration of APP vs WT; Column N: mean value of the fold change ratio of APP replicates vs reference replicate (126) - purple: up-regulated, green: down-regulated; Column O: mean value of the fold change ratio of WT replicates vs reference replicate (126), purple: up-regulated, green: down-regulated; Column P: Protein name/description; Column Q: Gene ontology information of every protein (XLSX 884 kb)
Rights and permissions
About this article
Cite this article
Mirzaei, M., Pushpitha, K., Deng, L. et al. Upregulation of Proteolytic Pathways and Altered Protein Biosynthesis Underlie Retinal Pathology in a Mouse Model of Alzheimer’s Disease. Mol Neurobiol 56, 6017–6034 (2019). https://doi.org/10.1007/s12035-019-1479-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-019-1479-4