Abstract
SARS-CoV-2 has a single-stranded RNA genome (+ssRNA), and synthesizes structural and non-structural proteins (nsps). All 16 nsp are synthesized from the ORF1a, and ORF1b regions associated with different life cycle preprocesses, including replication. The regions of ORF1a synthesizes nsp1 to 11, and ORF1b synthesizes nsp12 to 16. In this paper, we have predicted the secondary structure conformations, entropy & mountain plots, RNA secondary structure in a linear fashion, and 3D structure of nsp coding genes of the SARS-CoV-2 genome. We have also analyzed the A, T, G, C, A+T, and G+C contents, GC-profiling of these genes, showing the range of the GC content from 34.23 to 48.52%. We have observed that the GC-profile value of the nsp coding genomic regions was less (about 0.375) compared to the whole genome (about 0.38). Additionally, druggable pockets were identified from the secondary structure-guided 3D structural conformations. For secondary structure generation of all the nsp coding genes (nsp 1-16), we used a recent algorithm-based tool (deep learning-based) along with the conventional algorithms (centroid and MFE-based) to develop secondary structural conformations, and we found stem-loop, multi-branch loop, pseudoknot, and the bulge structural components, etc. The 3D model shows bound and unbound forms, branched structures, duplex structures, three-way junctions, four-way junctions, etc. Finally, we identified binding pockets of nsp coding genes which will help as a fundamental resource for future researchers to develop RNA-targeted therapeutics using the druggable genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
SARS-CoV-2 is an RNA virus of the Coronaviridae family, which is the causative pathogen of the current pandemic. The virus contains a single-stranded RNA (ssRNA) genome [1, 2] with approximately ~30 kb genomes that comprise one of the largest RNA among entire families of RNA viruses. The genome contains positive sense RNA, which includes several significant structural parts of the genome, such as 5′ cap structure and 3′ poly(A) tail [3, 4]. Moreover, the genome structure encodes about twenty-nine proteins [5]. Several researchers have tried to illustrate the genome structure. Manfredonia and Incarnato attempted to provide an insight view of the SARS-CoV-2 genome structure to offer strategies for developing new therapeutics. In this direction, they have illustrated different genome structural elements such as 5′ UTR structure, ribosomal FSE (frameshifting element), 3′ UTR structure [6]. Similarly, several structural elements were projected from the SARS-CoV-2 RNA genome, and in vivo structural characterization was performed for the genome. They have identified that 42 host proteins can interact with the different elements that were projected from the SARS-CoV-2 RNA. It might be helpful in developing new therapeutics against the virus [4]. Similarly, Cao et al. have developed the genome architecture of the SARS-CoV-2 RNA. The researchers have tried to understand the packing of the SARS-CoV-2 genome based on the RNA secondary structure. The study tried to comprehend the framework of the genome for the structure, function, and dynamics of the virus. Finally, they tried to design the siRNA for RNA-based drugs [7].
The large-scale characterization of RNA secondary structure might provide insight into the SARS-CoV-2 genome-wide organization. Simmonds illustrated a SARS-CoV-2 genome organization through RNA secondary structure and compared it with other Coronaviruses [8]. Several studies describe the significance of RNA secondary structures in the light of virus replication and their transcription and translation [9,10,11]. The genome of SARS-CoV-2 encodes two types of proteins: structural and non-structural proteins (nsps) (Fig. 1a). nsps are significant proteins that help the fundamental process of viruses such as transcription, envelope proteins production, and viral replication [12, 13]. All these NSPs support the RNA-dependent RNA polymerase (RdRp) to perform replication. Therefore, they act as replication machinery [14]. Scientists have used non-structural proteins (nsps) as drug targets [15]. NSP coding genes of SARS-CoV-2 are significant in this point of view. It has been noted that two open reading frames (ORF1a and ORF1b) are associated with the coding of 16 NSPs which are nsp1 to nsp16 (Fig. 1a) [16, 17]. ORF1a codes nsp1 to nsp11, and ORF1b codes nsp12 to nsp16 protein. The main protein includes nsp3 (papain-like proteinase protein), nsp5 (3C-like proteinase), nsp12 (RNA-dependent RNA polymerase, RdRp), nsp13 (helicase, HEL), NSP14 (exoRNAse), nsp15 (endoRNAse), nsp16 (2′-O-Ribose-Methyltransferase), and other non-structural proteins. Other significant nsp are nsp7 (cofactor for RNA-dependent RNA polymerase), nsp8 (cofactor for RdRp, primase, or 3′-terminal adenylyltransferase) [18]. Most of the research work has been performed to illustrate the structural and functional perspective of nsps (nsp1–16). However, less evidence illustrates insight into the nsp coding of genes (ORF1a and ORF1b) and their genomic structural arrangements. Therefore, it is urgent to understand the structural insight of the nsp coding genomic regions (ORF1a and ORF1b) and their structure-based genomic arrangements.
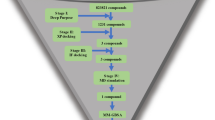
Schematic representation shows the single-stranded RNA (ssRNA) genome of SARS-CoV-2 and methodology flowchart of our study. a SARS-CoV-2 ssRNA genome contains non-structural protein-coding genes (nsp) (nsp1–to 16) and structural protein-coding genes. The nsp coding genes synthesized by ORF1a and ORF1b. ORF1a contains nsp1-to 11 protein-coding genes. ORF1b contains nsp12-to 16 protein-coding genes. b Schematic diagram shows a flowchart of the methodology of our work
RNA structure, both secondary and tertiary, plays critical functional roles in the life cycles of SARS-CoV-2. Using 3D structure, researchers are analyzing components of the structure that can help the structure-based drug discovery for this virus. Very few scientists have tried to illustrate the structure of the RNA genome of SARS-CoV-2. Rangan et al. have assessed RNA sequence conservation of the SARS-CoV-2 genome and found 106 RNA-based conserved regions that act as potential antiviral drug targets [19]. Romano et al. tried to understand the RNA structural-based replication machinery, where they found the interaction between the NSPs [20]. Sun et al. have analyzed RNA secondary structural elements of the SARS-CoV-2 RNA genome that plays a crucial role in viral infection. Their research might help to assess the antisense oligonucleotides targeting using RNA-based structural elements and will help in the therapeutics development against this virus [4]. Some scientists have contributed to identifying the RNA structure-based elements that can help therapeutic development. For example, Manfredonia et al. mapped the RNA structures throughout the SARS-CoV-2 genome. The RNA elements were identified from the genome. The secondary structure patterns have been generated to illustrate the SARS-CoV-2 genome. They have modeled the 3D structure of the RNA, which can help RNA-based therapeutic innovation [21]. However, the secondary and 3D structural RNA conformations of the nsp genes, their druggable genome, and the RNA-based therapeutic target in the virus genome are mostly unexplored. Therefore it is necessary to understand the in-depth analyses of the secondary, 3D RNA structural conformations of the SARS-CoV-2 genomes and the fundamental units of the druggable genome which can initiate the RNA targeting drug discovery against the virus.
In our work, we have tried to evaluate the secondary structure conformations, entropy and mountain plots, RNA secondary structure in a linear fashion, and 3D structure of NSP coding genes of the RNA genome of the SARS-CoV-2. We have predicted recent algorithms-based (deep learning-based) secondary structures, conventional algorithms-based (centroid and MFE-based) secondary structural conformations, entropy & mountain plots based on the secondary structures, single & overlapping structures of arc diagrams in a linear fashion of all the nsp coding genomic regions (nsp1–16). Finally, RNA 3D structural models have been predicted from those secondary structural conformations. Additionally, we have also analyzed the A, T, G, C, and A+T, G+C content, and GC-profiling to understand the GC content of all NSP coding genes. Finally, we have identified the secondary structure-guided druggable pockets (ligand binding sites) in RNA 3D structures conformation.
Method
Our analysis has been divided into three parts: secondary structure conformations analysis, 3D analysis, and the stability of the nsp coding genomic regions (nsp1–16). Several recent methods and current algorithms were used to analyze the RNA secondary structure, 3D structure, and stability of this genomic region.
Retrieval of the Genomic Sequence nsp Coding Genes of the SARS-CoV-2 Genome
We have retrieved the SARS-CoV-2 genome sequence and their nsp coding genes from NCBI (NCBI GenBank Id: MZ054889.1). The nsp coding genes and corresponding sequences’ position have been listed in Table 1. The nsp coding gene sequences (nsp1–16) were used for further analysis.
Secondary Structure Conformations Analysis
Deep Learning (DL) Based Secondary Structure Conformations Analysis
DL is an emerging approach exploring researchers in structural analysis from nucleic acids to proteins. We have used DL-based UFold server to analyze the DL-based secondary structure of RNA [22]. Using an encoder-decoder architecture, this server uses FCNs (Fully Convolutional Networks) to create fast and efficient RNA secondary structure. It uses the U-Net model for structure generation. The 17×L×L tensor is used as input for the U-Net model. The model predicted a score matrix Y that uses an L×L symmetric score for output through the contact map. In the case of traditional RNA secondary structure prediction methods, algorithms are mainly based on thermodynamic models through free energy minimization, which executes strong prior assumptions and slow speed to run. The UFold server proposes a deep learning-based method for the prediction of RNA secondary structure based directly on annotated data and nucleotide base-pairing rules. This approach might be more acceptable for computationally predicting the secondary structure of RNA [22].
Centroid-Based Secondary Structure Conformations Analysis
Another secondary structure conformation was predicted for the nsp coding genes based on centroid structure, used with minimum base-pair distance. Centroid-based secondary structures were predicted using an RNA sever (Vienna RNA website) [23]. From diverse types of structural RNAs, the centroid structure of RNA ensemble makes a 30.0% lesser prediction error which is measured by the positive predictive value. Moreover, it predicted the unavailable complete model that apparently comprises the unique native structure of studied RNA [24].
MFE-Based Secondary Structure Conformations Analysis
MFE-based secondary structures were generated for the nsp coding genomic regions. This structure was predicted by MFE-based secondary structure, utilizing a minimum of free energy. MFE structure was predicted using the Vienna RNA Website Server [23]. The server uses optimal computer folding employing external bases, the loop-based energy model unique decomposed loops. For predicting the MFE structure, alternatively, the RNA native structure of a given sequence is generated for each admissible base pair (isolated base pairs and stacking base pairs). The energetically best structure contains that base pair in the context of a minimum free energy landscape [25].
Entropy & Mountain Plots Analysis Based on the Secondary Structures
Entropy & mountain plots were generated based on the secondary structures. Entropy plots represent the thermodynamic assembly of a RNA secondary structure and the positional entropy of a RNA secondary structure. Entropy plots were predicted using the Vienna RNA website server [23]. The entropy and mountain plot are exclusively useful for long nucleotide sequences where conventional structure illustrations become a disorder. It is an axis (xy-diagram) oriented plotting, where the number of base pairs circling a sequence position versus the nucleotide position. The sets of computer program transforms a dot plot into the mountain plot coordinates which the graphical user interface can visualize. The resulting plot displays three significant curves, two mountain plots derived from the MFE structure (red), a positional entropy curve (green), and the pairing probabilities (black) [25].
Mountain plots were generated using the same server. It represents the secondary structure through a height and informs us of the positional height of an RNA secondary structure. The height m(k) is allotted by the number of base pairs included in the base at position k.
Arc Diagrams to Understand the Secondary Structure Conformations Linearly Both in a Single Manner, Overlapping Manner, and Trans-RNA–RNA Interactions
The Arc diagrams have been developed through the R-chie server [26]. Arc diagrams were used to describe the secondary structure conformations linearly, both in a single & overlapping manner. To develop the Arc diagrams, the server used the helix prediction method TRANSAT (TRANSAT algorithm), which is visualized through the R package and graphically represented through the line diagram using the package (R4RNA). Finally, the result will help us to understand trans-RNA–RNA interactions. Using the server, we have illustrated the trans-RNA–RNA interactions [27].
3D Structural Conformation Analysis
3D Structural Conformation Based on Line Chain Model And Wire Model
Predicting the 3D structure of RNA and its proper understanding remains a major challenge. The 3D structure can help components of RNA structures and assist in positioning the helical regions of RNA. 3D structure conformation of nsp coding genomic regions (nsp1–16) was developed by RNAComposer [28, 29]. Here we developed two types of 3D models: the line chain 3D model and the wire 3D model. The RNAComposer server is a user-friendly and freely accessible server target for the 3D structure prediction of RNA (up to 500 nucleotides). It employs fully automated fragment assembly-based methods on the RNA secondary structure detailed by the user. Presently, such novel computational methods based on the incorporation of distance restraints resultant strategies are emerging in the field of computational biology [29].
Identification of Druggable Pockets (Ligand Binding Sites) in RNA 3D Structures Conformation and Its Comparison with the Secondary Structure
The identification of druggable pockets is an interesting area of structure-based drug design [30]. Identification of druggable pockets in RNA 3D structural conformation in the nsp coding genes using Voronoia 4-ever server [31]. The server calculates ligand-binding pockets, solvent-free surfaces, packing defects, etc. It also calculates the packing density for every atom packing density is denoted by PD, which is calculated using the following formula:
Here, using the Voronoi cell method, VvdW is the volume allocated to each atom, which is just inside the vdW field of the atom. Here, Vse is defined as the remaining volume, and it is excluded from the solvent.
GC Content and GC-Profile Analysis Throughout the nsp Coding Genome
Stability Analysis Through A, T, G, C, and A+T, G+C Content
Scientists have tried to analyze the GC-rich region of the genome of the virus [32]. It can help to understand the GC content bias regions. We used the GC content calculator and calculated A, T, G, C, and A+T, G+C content of the nsp coding genomic regions (nsp1–16) SARS-CoV-2 genome using the retrieved genome sequence.
GC-Profiling to Understand the Content
We have analyzed GC-profiling of the whole genome of SARS-CoV-2 through a GC-profiling plot through the GC-Profile server. We have evaluated GC-profiling of the nsp coding genomic regions of the virus through GC-profiling plot through the GC-Profile server and finally compared the GC-profile of both the regions (the nsp coding genomic regions and whole genome). GC-Profile server uses a segmentation algorithm to estimate the GC-Profile. In the segmentation algorithm, the genome order index is calculated, which is denoted as S. The S is defined as follows:
GC-Profile is a web-based, interactive software system mainly used to analyze eukaryotic and prokaryotic genomes. It offers a qualitative as well as a quantitative view of organism genome organization. Server-based results also reproduce the relationships among the G+C content and other significant genomic landscapes, e.g., distributions of CpG islands and genes, which are also analyzed comprehensibly. This proved that GC-profile is an appropriate early point for analyzing the isochore structure of higher eukaryotic genomes and identifying genomic islands in prokaryotic genomes [33].
Finally, the server presented a cumulative GC-profile of the genome or the nsp coding genomic regions.
The entire methodology of our work has been described through a flowchart (Fig. 1b).
Result
Secondary Structure Conformations
Deep Learning (DL)-Based Secondary Structure Conformations
Deep learning (DL)-based secondary structures were developed coding regions of the different nsp (nsp1–16) of the SARS-CoV-2 genome. It illustrated the DL-based secondary structures ORF1a and ORF1b regions. The DL-based secondary structures of ORF1a were illustrated through the coding genomic regions of nsp1-11 (Fig. 2A1–A11). Similarly, DL-based secondary structures of ORF1b were illustrated through the coding genomic regions of nsp12-16 (Fig. 2B1–B5). The nsp1 coding region shows more than 14 internal loops. We also found 2 to 3 larger internal loops (Fig. 2A1). In this region, long stems are found compared to other regions. Similarly, the nsp2 coding region shows more than 14 internal loops (Fig. 2A2). We also found 2 to 3 large internal loops. The bulge loop, multi-branched, stem, hairpin loop, and internal loop were also noticed in this region. Similarly, the nsp3 coding region shows more number of multi-branched loops. We also found the bulge loop, stem, hairpin loop, and internal loop in this region (Fig. 2A3). The nsp4 coding region illustrates three larger multi-branched loops and several small multi-branched loops (Fig. 2A4). Again, the nsp5 coding region illustrates one larger multi-branched loop in conjunction with several small multi-branched loops (Fig. 2A5). nsp6 coding region illustrates one larger multi-branched loop.along with four medium-sized multi-branched loops other several small multi-branched loops (Fig. 2A6). Also, we found one larger multi-branched loop.along with two medium-sized multi-branched loops in the nsp7 coding region (Fig. 2A7). In the nsp8 coding region, a large multi-branched loop along with a bulge loop, stem, hairpin loop, and internal loop was noted (Fig. 2A8). Similarly, in the nsp9 coding region, we found one larger multi-branched loop along with a bulge loop, stem, hairpin loop, and internal loop (Fig. 2A9). In the nsp10 coding region, we found bigger multi-branched loops which the structure complex (Fig. 2A10). However, the nsp11 coding region’s structure is simpler than other regions that show a bulge loop, internal loop, and stem (Fig. 2A11). In the ORF1b regions contain, nsp12 coding region is more complex compared to other regions where we noted three multi-branched loops (Fig. 2B1). Similarly, in the nsp13 coding region, we found several internal loops along with multi-branched loops (Fig. 2B2). Again, the nsp14 coding region shows five multi-branched loops along with a bulge loop, hairpin loop, and internal loop (Fig. 2B3). Similarly, the nsp15 coding region shows three multi-branched loops along with other structures such as the bulge loop, hairpin loop, and internal loop (Fig. 2B4). Finally, we found two multi-branched loops along with other structures, such as the internal loop, hairpin loop, and the bulge in the nsp15 coding region (Fig. 2B5).
RNA secondary structure model predicted by deep learning (DL) of all nsp coding regions (nsp1–nsp16) which shows the different components of the structure such as stem-loop (SL), multi-branch loop, pseudoknot, and bulge etc. A1 Secondary structure conformations of nsp1 coding gene and their components. A2 Secondary structure conformations of nsp2 coding gene and their components. A3 Secondary structure conformations of nsp3 coding gene. A4 Secondary structure conformations of nsp4 coding gene and their components. A5 Secondary structure conformations of nsp5 coding gene and their components. A6 Secondary structure conformations of nsp6 coding gene and their components. A7 Secondary structure conformations of nsp7 coding gene and their components. A8 Secondary structure conformations of nsp8 coding gene and their components. A9 Secondary structure conformations of nsp9 coding gene and their components. A10 Secondary structure conformations of nsp10 coding gene and their components. A11 Secondary structure conformations of nsp11 coding gene and their components. B1 Secondary structure conformations of nsp12 coding gene and their components. B2 Secondary structure conformations of nsp13 coding gene and their components. B3 Secondary structure conformations of nsp14 coding gene and their components. B4 Secondary structure conformations of nsp15 coding gene. B5 Secondary structure conformations of nsp16 coding gene and their components
Centroid-Based Secondary Structure Conformations
Like the DL-based secondary structures, we also developed centroid-based secondary structure conformations of coding regions for the nsps (nsp1–16) of the SARS-CoV-2 genome. We developed centroid-based secondary structures of ORF1a, which were illustrated through the coding genomic regions of nsp1-11 (Fig. S1A1–A11). Similarly, the centroid-based secondary structures of ORF1b were demonstrated through the coding genomic regions of the nsp12-16 (Fig. S1B1–B5). In both cases (ORF1a and ORF1b), the structure of the coding genomic regions of the nsp1-16 are more simple compared to DL-based secondary structures. The nsp1 shows three multi-branched loop structures (Fig. S1A1). Similarly, nsp2 shows three multi-branched loop structures (Fig. S1A2). Likewise, nsp3 shows a linear-type structure, and several stem and loop structures have been generated from the linear structure (Fig. S1A3). Similarly, in the nsp4, three multi-branched loops with a bigger loop structure are noted (Fig. S1A4). In the nsp5, five to six multi-branched loops are observed with a different bulge. We also found hairpin loops and internal loops in the structure (Fig. S1A5). In the nsp6, a major multi-branched loop is observed (Fig. S1A6). Similarly, in the nsp7, two major multi-branched loops are observed (Fig. S1A7). Also, in the nsp8, we noted three major multi-branched loops along with several hairpin loops and internal loops in the entire structure (Fig. S1A8). Likewise, in the nsp9, two major multi-branched loops are noted, and one structure is noted with a bulge (Fig. S1A9). At the same time, several stems, hairpin loops, and internal loops were found in the nsp10 in conjunction with two multi-branched loops (Fig. S1A10). The nsp11 coding region formed a small structure due to its small sequence length (Fig. S1A11). Likewise, in the nsp12, about nine multi-branched loops are noted in the structure of the region. We also found several hairpin loops and internal loops in this region (Fig. S1B1). Similarly, two significant multi-branched loops are noted, which are associated together in the nsp13 (Fig. S1B2). The nsp14 developed a linear-type structure with six multi-branched loops (Fig. S1B3). Likewise, nsp15 contains four multi-branched loops, and the middle multi-branched loop encloses two major bulge structures (Fig. S1B4). Finally, it has been noted that nsp16 formed two multi-branched loops, and the last multi-branched loop encloses one major bulge structure. The first one contains several stems, loops, and hairpin structures (Fig. S1B5).
MFE-Based Secondary Structure Conformations
Like the DL-based and centroid-based secondary structures, we also developed MFE-based conformations of coding regions of the nsps (nsp1–16) of the genome of the virus. We developed MFE-based secondary structures of ORF1a with the coding genomic regions of nsp1-11 (Fig. S2A1–A11). Likewise, the ORF1b was demonstrated through the MFE-based conformations of the coding genomic regions of the nsp12-16 (Fig. S2B1–B5). The nsp1 shows two main multi-branched loops (Fig. S2A1). Similarly, nsp2 shows six multi-branched loop structures (Fig. S2A2). The nsp3 depicts a linear-type structure with several stems and loop structures (Fig. S2A3). This MFE-based secondary of nsp3 is similar to the centroid-based secondary structure. Similarly, It has been noted aboutgenome identifies host proteins vulnerable five multi-branched loops in the nsp4 (Fig. S2A4). Five to six multi-branched loops are observed, a similar type of centroid-based secondary structure (Fig. S2A5). In the nsp6, we have also observed five to six multi-branched loops (Fig. S2A6). Likewise, in the nsp7, one major multi-branched loop is observed. We noted the bulge in the structure (Fig. S2A7). Similarly, in the nsp8, we noted three major multi-branched loops with different components of RNA secondary structure. We noted several structural components (stem, loops, hairpin loop) in the first and last multi-branched structure (Fig S2A8). At the same time, two major multi-branched loops are noted in the nsp9, and the last structure is noted with different structural components (Fig. S2A9). At the same time, we found two multi-branched loops in the nsp10 (Fig. 4A10). Similarly, we found a linear structure that contains about eight multi-branched loops in the structure of the region (Fig. S2B1). In the same way, we found a linear structure of the nsp13 (Fig. S2B2). At the same time, we noted that nsp14 had created a linear-type structure with five multi-branched loops (Fig. S2B3). It has been noted that nsp15 contains about three multi-branched loops, and the middle and last multi-branched loop encloses several stems, loops, and hairpin loop structures (Fig. S2B4). At last, we found that nsp16 formed two multi-branched loops, and the multi-branched loop contained several stems, loops, and hairpin structures (Fig. S2B5).
Entropy and Mountain Plots Based on The Secondary Structures
We depicted the entropy and mountain plots of all nsp coding regions (Fig. S3A1–A11, B1–B5). The nsp1 shows entropy and mountain plots. Maximum entropy exhibited 2.0 (in between 300 and 500 locations), and the height is about 40 in the region of nsp1 (Fig. S3A1). Similarly, nsp2 shows.maximum entropy of 2.0 in between the 1 to 500 location, and the height is about 100 (Fig. S3A2). The nsp3 illustrates a maximum entropy of 2.0 (in between 1 and 1000 locations) and a maximum height of 200 (Fig. S3A3). Similarly, in the nsp12, we noted the energy peaks with a value of 3 (between 1000–1500 and 2000–2500 locations). We noted several energy peaks in those regions. The mountain plot shows a maximum height of about 120, located in the first part (Fig. S3B1). In the region of nsp15, we noted maximum entropy, and it was exhibited at 3.0 (near about 1000 location), and the height of the mountain plots is about 60 to 90 (Fig. S3B4). Again, in the region of nsp16, we noted maximum entropy, and it was exhibited 2.0 to 2.5 (in between 200–400 location, and near about 600) and the height of the mountain plots is about 40 to 60 (Fig. S3B5).
Arc Diagrams of the Secondary Structure Conformations Linearly Both in the Single Manner, Overlapping Manner, and Trans-RNA–RNA Interactions
The arc diagram illustrates the probable base pair of the RNA structure which connects the relevant RNA sequence to the secondary structure. It is depicted as a horizontal line from left to right (5′ to 3′ ends).
We depicted the Arc diagrams of the secondary structure conformations linearly both in the single and overlapping manner for all nsp coding regions (nsp1–16) (Fig. 3A1–A11, B1–B5). Relating to the TRANSAT algorithm, the nsp1 coding region shows a high base paring with a value of [1e-06, 1e-05] in the case of a single line. Similarly, it also shows the high possibility of base pairing in the case of an overlapping manner. More linear arc structures were found in the coding region of nsp1, nsp3, nsp4, nsp7, nsp9, nsp10 in the ORF1a (Fig. 3A1, A3, A4, A7, A9, A10). Similarly, we found more linear arc structures in nsp12 and nsp15 in the ORF1b (Fig. 3B1, B4). It has been noted that the coding region of nsp7 developed denser linear arc structures in both cases of single, overlapping fashion among all of the nsps of ORF1b and ORF1b.
Arc diagrams of the secondary structure conformations linearly both in the single manner, overlapping manner of all nsp coding regions (nsp1–nsp16). A1 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp1 coding gene. A2 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp2 coding gene. A3 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp3 coding gene. A4 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp4 coding gene. A5 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp5 coding gene. A6 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp6 coding gene. A7 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp7 coding gene. A8 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp8 coding gene. A9 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp9 coding gene. A10 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp10 coding gene. A11 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp11 coding gene. B1 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp12 coding gene. B2 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp13 coding gene. B3 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp14 coding gene. B4 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp15 coding gene. B5 Arc diagrams of the secondary structure conformations in the single manner, overlapping manner of nsp16 coding gene
3D Structural Conformation Analysis
3D structural Conformation Based on Line Chain Model and Wire Model
We developed the 3D model of the secondary structure to understand the folding for all the nsp coding regions (nsp1–16) (Fig. 4A1–A11, B1–B5). We have understood the proper folding of the RNA structure in 3D structure space. Here we have identified the stem and loop structure in the line chain and wire model for all the nsp coding regions. In the ORF1a region, it has been found that the nsp7 coding region contains more stem structures (Fig. 4A7). At the same time, in ORF1b regions, we found that the nsp13 coding region contains more stem structures (Fig. 4B2).
3D model of the RNA of nsp coding regions (nsp1–nsp16) shows the different structural components in the line chain model and wire model. A1 3D model (line chain and wire model) of nsp1 coding gene. A2 3D model of nsp2 coding gene. A3 3D model (line chain and wire model) of nsp3 coding gene. A4 3D model (line chain and wire model) of nsp4 coding gene. A5 3D model (line chain and wire model) of nsp5 coding gene. A6 3D model of nsp6 coding gene. A7 3D model (line chain and wire model) of nsp7 coding gene. A8 3D model (line chain and wire model) of nsp8 coding gene. A9 3D model (line chain and wire model) of nsp9 coding gene. A10 3D model (line chain and wire model) of nsp10 coding gene. A11 3D model (line chain and wire model) of nsp11 coding gene. B1 3D model (line chain and wire model) of nsp12 coding gene. B2 3D model (line chain and wire model) of nsp13 coding gene. B3 3D model (line chain and wire model) of nsp14 coding gene. B4 3D model (line chain and wire model) of nsp15 coding gene. B5 3D model of nsp16 coding gene
Presently, understanding the sequence-related RNA helix conformation is an interesting area. Researchers are also trying to understand the RNA helices pattern of the 3D model of RNA structure [34]. Several scientists are trying to assess the bound and unbound form in the RNA 3D model [35]. RNA 3D model of different bound and unbound forms has been shown in the case of base pairing within RNA helices of this conformation. We have also understood the packing of RNA in 3D conformation. Here, we identified the duplex, branched structure, three-way junction, and four-way junction all the nsp coding regions (nsp1–16).
Identification of Druggable Pockets (Ligand Binding Sites) in RNA 3D Structures Conformation and Its Comparison with the Secondary Structure
Finally, we identified the drug-binding pocket of all the nsp coding regions (nsp1–16). Here, we have represented the three nsp coding structures (nsp7, nsp12, and nsp13) (Fig. 5a–c). In all cases, we have shown two drug-binding pockets in each RNA 3D structures conformation. Each drug-binding pocket in each RNA 3D structure has been compared with the secondary structure of RNA predicted from the nsp coding structures.
Identified ligand-binding pocket in RNA 3D structures (nsp coding) conformation guided with the secondary structure. a Identified ligand-binding pocket in nsp7 coding RNA 3D structures conformation guided with the secondary structure. b Identified ligand-binding pocket in nsp12 coding RNA 3D structures conformation guided with the secondary structure. c Identified ligand-binding pocket in nsp13 coding RNA 3D structures conformation guided with the secondary structure. In every case, we have shown two binding pockets
These three nsp regions (nsp7, nsp12, and nsp13) code three significant proteins which are cofactor for RNA-dependent RNA polymerase (nsp7), RNA-dependent RNA polymerase, or RdRp (nsp12), helicase or HEL (nsp13). All these proteins are associated with the viral life cycle. Therefore, druggable pockets (ligand binding sites) will help identify the drug candidates targeting these regions, affecting the viral life cycle. siRNA-based molecules can be predicted using those druggable pockets. Previously, we identified siRNA-based therapeutic candidates targeting RdRp [36].
GC Content and GC-Profile Analysis Throughout the nsp Coding Genome
A, T, G, C, and A+T, G+C Content of the nsp Coding Genomic Regions (nsp1–16)
Here, we have calculated the A, T, G, C, and A+T, G+C content of the nsp coding genomic regions from their sequence space GC-profiling to understand the content of the nsp coding genomic regions (nsp1–16) (Fig. S4A1–A11 and Fig. S4B1–B5). We found that A+T is higher in all the nsp coding regions than G+C content. The G+C content of all the nsps (nsp1 to nsp11) of ORF1a are noted as 48.52%, 39.76%, 35.75%, 36.53%, 38.13%, 36.21%,37.39%, 38.22%, 40.12%, 42.45%, 53.80%. At the same time, G+C content of all the nsps (nsp12 to nsp16) of ORF1b are noted as 37.18%, 38.21%, 38.08%, 34.23%, 35.46%.
GC-Profiling of the nsp Coding Genomic Regions (nsp1–16) and Comparison with the Whole Genome of SARS-CoV-2
We analyzed the GC-profiling of the nsp coding regions of ORF1a and ORF1b. The graph shows the highest peak at 4000 bp and 20,000 bp regions. At the same time, the graph shows the lowest peak, in between 9000 to 12,000 bp regions (Fig. 6a).
GC-profiling of the nsp coding genomic regions a GC-profiling of both the nsp coding regions of ORF1a and ORF1b. It mapped the GC contents in every region of the nsp coding genes of ORF1a and ORF1b regions. b GC-profiling of the whole genome. However, no segmentation in the GC contents was noted in both cases
The GC-profiling of the whole genome shows the highest peak of 4000 bp and 20,000 bp regions, and suddenly, a very high peak was noted in the last part of the genome. At the same time, the graph shows the two lowest peaks, at 9000 to 12,000 bp and around 23,000 bp regions (Fig. 6b).
However, it has been observed that the GC-profile value of the nsp coding genomic regions (nsp1–16) is less (about 0.375) compared to the whole genome (about 0.38).
Discussion
Presently, researchers are trying to understand the architecture of RNA, especially the RNA structure in the genome [7, 37]. RNA structure can be utilized in different other conditions, such as the different significant scientific events (host immunity and the evolution of virus) [38]. At the same time, an understanding of architecture in RNA structures in secondary structural form and 3D structural form is important to illustrate the replication process [14, 39]. Simultaneously, researchers are trying to understand the druggable genome, one of the focus areas for structure-based drug design. Therefore, RNA structure in the genome component will help with RNA-targeted drug design [40,41,]. In this direction, scientists are trying to solve the RNA architecture in the SARS-CoV-2 genome. Understanding the RNA secondary and 3D structure of the SARS-CoV-2-ssRNA genome is significant in this pandemic scenario and will help the RNA-targeted therapeutic design. Manfredonia et al. have tried to evaluate the structural signature of the SARS Cov-2 genome and identified RNA structural components with therapeutic significance [21]. In this direction, we have illustrated the secondary structure, 3D structure rated, and GC-profile of the nsp coding regions of the SARS-CoV-2 genome. Finally, we have identified ligand-binding pockets of the nsp coding regions. Therefore, our study is very significant and will initiate the discovery of RNA-based therapeutics against SARS-CoV-2.
Druggable genome-wide therapeutic siRNA discovery is one of the effective approaches for developing antisense oligonucleotide therapeutics [41,42,43]. Cao et al. have studied the SARS-CoV-2 RNA genome architecture and siRNA-based therapeutic design. They have identified four observed siRNAs, and no infection was found in the cell after treatment of siRNAs [7]. Sun et al. illustrated the RNA genome architecture of the SARS-CoV-2 and further explored siRNA-based therapeutic design. In this study, researchers have tried to identify the different genomic regions that are potential targets for siRNA. The study will help to develop siRNA-based therapies for SARS-CoV-2 [4]. Previously, we have also identified siRNAs targeting the mRNA of RdRp, an example of the RNA-based therapeutic discovery for SARS-CoV-2. However, we have targeted the mRNA of RdRp of SARS-CoV-2. Several researchers have tried to understand the transcriptomic profile of SARS-CoV-2 [37]. It will help to comprehend the SARS-CoV-2 life cycle and its pathogenicity. At the same time, the study will also help to explore RNA-based therapeutic design.
The druggability characterization within the SARS-CoV-2 proteome is a significant effort for viral drug target-based therapy. Cavasotto et al. [44] measured the functional role of each SARS-CoV-2 protein encoded from the whole RNA genome. Subsequently, they analyzed the main structural landscapes, using the experimental homology models or structural data using in-depth in silico druggability assessment of the viral proteome [45].
Deep learning-based algorithms and tools are important to illustrate the genome structure [46] and are used regularly for RNA secondary structure generation [47]. Sun et al. illustrated the RNA genome architecture of the SARS-CoV-2 using a deep learning tool and developed the RNA secondary structure of this virus. Along with the conventional method, our study used the deep learning-based modern tool to develop the secondary structure of the nsp coding regions of the SARS-CoV-2 RNA genome. The study compared the predicted deep learning-based secondary structures and conventional secondary structures (Centroid and MFE-based) (Table 2).
Simultaneously, we calculated the GC content of the nsp coding regions. We found that the range of the GC content was 34.23% to 48.52%. Although, we found the nsp11 coding region is about 53.80%. However, the sequence length of this part is too short, so, we have not considered this region. Again, we evaluated the GC-profile of the nsp coding regions and compared to the whole SARS CoV-2 genome. Several other researchers have calculated the GC usage of SARS-CoV-2. Li et al. calculated the GC usage of the genes of SARS-CoV-2 and found lower GC content of the genome. It has been reported that the GC content of the virus coding sequence is around 38%. At the same time, researchers found that the GC content of the vertebrates along with the humans is around 60% [48].
Additionally, our study has identified drug-binding pockets from the RNA genome, and it is a promising approach for the therapeutic development of SARS-CoV-2 or other infective RNA viruses. Therefore, our work has immense value.
Limitations
Several researchers have used the next-generation platform (such as SHAPE-MaP evaluation) to illustrate the architecture of the RNA genome. Due to the lack of resources, we were unable to find the next-generation platform to develop the genome architecture of this RNA virus. However, to fulfill the limitation, we have used modern algorithms (deep learning-based algorithms) to illustrate the secondary structure of the genome structure. We have modeled the 3D structure of all nsp coding genes due to the lack of the crystal or cryo-electron structure. However, we urge researchers to develop crystal or cryo-electron structures of all the nsp coding genes.
The information-centric terminology in biological systems can be considered properly when it is embedded in theoretical biology, and it validates bioinformatics-based analysis. Successive models and experimental validation is exclusively required for any scientific experiment. Many researchers utilizing artificial intelligence are currently exploring new illustrations of information processing systems, often inspired by biological systems, e.g., protein-nucleotide network models and biomolecule pattern recognition. Our study emphasized advanced modeling practices of nsp coding genomic regions of SARS-CoV-2-ssRNA to understand and develop RNA-targeted therapeutics using the druggable genome.
Although all sorts of modeling, structure, and pattern analysis were considered distinct endeavors, we felt that for bioinformatics research, they were both desired and should be united to compare the outcome of such models with “real” data [49, 50].
Future Prospective
Our data illustrated nsp coding regions of the SARS-COV-2 genome. The data of our work act as the fundamental resources to unfold more about the SARS-CoV-2 life cycle, replication, and unknown architecture of the genome. At the same time, understanding the secondary and 3D structure of the genome and its composition is important. However, the secondary structure-guided 3D structure and its identified ligand-binding pocket will shed light on the druggable genome for the researcher to develop multiple therapeutics against the virus.
Conclusion
Our work not only emphasized the secondary and 3D structure development but also identified all the elements of secondary structure, such as stem-loop structure, multi-branch loop, pseudoknot structure, and the bulge structural components in the secondary structure in the nsp coding genes. We also identified the unbound structure, branched structure, duplex structure, three-way junction, four-way junction, etc., in the 3D structure. The study will help to identify the structural elements of the SARS-COV-2 genome.
Our data of binding pocket identification will offer a primary platform for RNA-targeted therapeutic development. It will help identify the druggable genome and unique RNA-based therapeutic target, which will help structure-based drug design against SARS-CoV-2. Finally, the identified binding pocket in the nsp coding regions will help future researchers to multiple small molecule-based ligand discoveries and antisense oligonucleotide therapeutics discoveries targeting those genomic regions to fight against the virus.
Data Availability
The authors confirm that the data supporting the findings of this study are available as Supplementary figures.
References
Umair, M., Ikram, A., Salman, M., Khurshid, A., Alam, M., Badar, N., Suleman, R., Tahir, F., Sharif, S., Montgomery, J., & Whitmer, S. (2021). Whole-genome sequencing of SARS-CoV-2 reveals the detection of G614 variant in Pakistan. PLoS ONE, 16(3), e0248371.
De Wit, E., Van Doremalen, N., Falzarano, D., & Munster, V. J. (2016). SARS and MERS: Recent insights into emerging coronaviruses. Nature Reviews Microbiology, 14(8), 523–534.
Wu, F., Zhao, S., Yu, B., Chen, Y. M., Wang, W., Song, Z. G., Hu, Y., Tao, Z.W., Tian, J. H., Pei, Y. Y., & Yuan, M. L. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269.
Sun, L., Li, P., Ju, X., Rao, J., Huang, W., Ren, L., Zhang, S., Xiong, T., Xu, K., Zhou, X., & Gong, M. (2021). In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs. Cell, 184(7), 1865–1883.e20.
Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., O’Meara, M. J., Rezelj, V. V., Guo, J. Z., Swaney, D. L., & Tummino, T. A. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature, 583(7816), 459–468.
Manfredonia, I., & Incarnato, D. (2021). Structure and regulation of coronavirus genomes: State-of-the-art and novel insights from SARS-CoV-2 studies. Biochemical Society Transactions, 49(1), 341–352.
Cao, C., Cai, Z., Xiao, X., Rao, J., Chen, J., Hu, N., Yang, M., Xing, X., Wang, Y., Li, M,. & Zhou, B. (2021). The architecture of the SARS-CoV-2 RNA genome inside virion. Nature Communications, 12(1), 1–14.
Simmonds, P. (2020). Pervasive RNA secondary structure in the genomes of SARS-CoV-2 and other coronaviruses. MBio, 11(6), e01661-e1720.
Züst, R., Miller, T. B., Goebel, S. J., Thiel, V., & Masters, P. S. (2008). Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. Journal of Virology, 82(3), 1214–1228.
Yang, D., & Leibowitz, J. L. (2015). The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Research, 206, 120–133.
Zhang, Y., Huang, K., Xie, D., Lau, J. Y., Shen, W., Li, P., Wang, D., Zou, Z., Shi, S., Ren, H., & Wang, Y. (2021). In vivo structure and dynamics of the SARS-CoV-2 RNA genome. Nature Communications, 12(1), 1–12.
Mousavizadeh, L., & Ghasemi, S. (2021). Genotype and phenotype of COVID-19: Their roles in pathogenesis. Journal of Microbiology, Immunology and Infection, 54(2), 159–163.
Mariano, G., Farthing, R. J., Lale-Farjat, S. L., & Bergeron, J. R. (2020). Structural characterization of SARS-CoV-2: Where we are, and where we need to be. Frontiers in Molecular Biosciences, 7, 344.
Malone, B., Urakova, N., Snijder, E. J., & Campbell, E. A. (2022). Structures and functions of coronavirus replication–transcription complexes and their relevance for SARS-CoV-2 drug design. Nature Reviews Molecular Cell Biology, 23(1), 21–39.
Raj, R. (2021). Analysis of non-structural proteins, NSPs of SARS-CoV-2 as targets for computational drug designing. Biochemistry and Biophysics Reports, 25, 100847.
Rahimi, A., Mirzazadeh, A., & Tavakolpour, S. (2021). Genetics and genomics of SARS-CoV-2: A review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics, 113(1), 1221–1232.
Subissi, L., Imbert, I., Ferron, F., Collet, A., Coutard, B., Decroly, E., & Canard, B. (2014). SARS-CoV ORF1b-encoded non-structural proteins 12–16: Replicative enzymes as antiviral targets. Antiviral Research, 101, 122–130.
V’kovski, P., Kratzel, A., Steiner, S., Stalder, H., & Thiel, V. (2021). Coronavirus biology and replication: implications for SARS-CoV-2. Nature Reviews Microbiology, 19(3), 155–170.
Rangan, R., Zheludev, I. N., Hagey, R. J., Pham, E. A., Wayment-Steele, H. K., Glenn, J. S., & Das, R. (2020). RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: A first look. RNA, 26(8), 937–959.
Romano, M., Ruggiero, A., Squeglia, F., Maga, G., & Berisio, R. (2020). A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells, 9(5), 1267.
Manfredonia, I., Nithin, C., Ponce-Salvatierra, A., Ghosh, P., Wirecki, T. K., Marinus, T., Ogando, N. S., Snijder, E. J., van Hemert, M. J., Bujnicki, J. M., & Incarnato, D. (2020). Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Research, 48(22), 12436–12452.
Fu, L., Cao, Y., Wu, J., Peng, Q., Nie, Q., & Xie, X. (2022). Ufold: Fast and accurate RNA secondary structure prediction with deep learning. Nucleic Acids Research, 50(3), e14–e14.
Gruber, A. R., Lorenz, R., Bernhart, S. H., Neuböck, R., & Hofacker, I. L. (2008). The vienna RNA websuite. Nucleic Acids Research, 36(suppl_2), W70–W74.
Ding, Y., Chan, C. Y., & Lawrence, C. E. (2005). RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA, 11(8), 1157–1166. https://doi.org/10.1261/rna.2500605
Li, Y., & Zhang, S. (2011). Finding stable local optimal RNA secondary structures. Bioinformatics, 27(21), 2994–3001. https://doi.org/10.1093/bioinformatics/btr510
Lai, D., Proctor, J. R., Zhu, J. Y. A., & Meyer, I. M. (2012). R-CHIE: A web server and R package for visualizing RNA secondary structures. Nucleic Acids Research, 40(12), e95–e95.
Tsybulskyi, V., Mounir, M., & Meyer, I. M. (2020). R-chie: A web server and R package for visualizing cis and trans RNA–RNA, RNA–DNA and DNA–DNA interactions. Nucleic Acids Research, 48(18), e105–e105.
Biesiada, M., Purzycka, K. J., Szachniuk, M., Blazewicz, J., & Adamiak, R. W. (2016). Automated RNA 3D structure prediction with RNAComposer. RNA Structure Determination (pp. 199–215). New York, NY: Springer.
Biesiada, M., Pachulska-Wieczorek, K., Adamiak, R. W., & Purzycka, K. J. (2016). RNAComposer and RNA 3D structure prediction for nanotechnology. Methods, 103, 120–127. https://doi.org/10.1016/j.ymeth.2016.03.010
Pérot, S., Sperandio, O., Miteva, M. A., Camproux, A. C., & Villoutreix, B. O. (2010). Druggable pockets and binding site centric chemical space: a paradigm shift in drug discovery. Drug Discovery Today, 15(15–16), 656–667.
Staritzbichler, R., Ristic, N., Goede, A., Preissner, R., & Hildebrand, P. W. (2021). Voronoia 4-ever. Nucleic Acids Research, 49(W1), W685–W690.
Miyata, H., Tsunoda, H., Kazi, A., Yamada, A., Khan, M. A., Murakami, J., Kamahora, T., Shiraki, K., & Hino, S. (1999). Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. Journal of Virology, 73(5), 3582–3586.
Gao, F., & Zhang, C. T. (2006). GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Research, 34(Web Server), W686–W691. https://doi.org/10.1093/nar/gkl040
Yesselman, J. D., Denny, S. K., Bisaria, N., Herschlag, D., Greenleaf, W. J., & Das, R. (2019). Sequence-dependent RNA helix conformational preferences predictably impact tertiary structure formation. Proceedings of the National Academy of Sciences, 116(34), 16847–16855.
Abraham, M., Dror, O., Nussinov, R., & Wolfson, H. J. (2008). Analysis and classification of RNA tertiary structures. RNA, 14(11), 2274–2289.
Shawan, M. M. A. K., Sharma, A. R., Bhattacharya, M., Mallik, B., Akhter, F., Shakil, M. S., Hossain, M. M., Banik, S., Lee, S. S., Hasan, M. A., & Chakraborty, C. (2021). Designing an effective therapeutic siRNA to silence RdRp gene of SARS-CoV-2. Infection, Genetics and Evolution, 93, 104951.
Kim, D., Lee, J. Y., Yang, J. S., Kim, J. W., Kim, V. N., & Chang, H. (2020). The architecture of SARS-CoV-2 transcriptome. Cell, 181(4), 914-921.e10.
Smyth, R. P., Negroni, M., Lever, A. M., Mak, J., & Kenyon, J. C. (2018). RNA structure—A neglected puppet master for the evolution of virus and host immunity. Frontiers in Immunology., 9, 2097.
Hillen, H. S., Kokic, G., Farnung, L., Dienemann, C., Tegunov, D., & Cramer, P. (2020). Structure of replicating SARS-CoV-2 polymerase. Nature, 584(7819), 154–156.
Warner, K. D., Hajdin, C. E., & Weeks, K. M. (2018). Principles for targeting RNA with drug-like small molecules. Nature Reviews Drug Discovery, 17(8), 547–558.
Scott, H. L., Buckner, N., Fernandez-Albert, F., Pedone, E., Postiglione, L., Shi, G., Allen, N., Wong, L. F., Magini, L., Marucci, L., & O’Sullivan, G. A. (2020). A dual druggable genome-wide siRNA and compound library screening approach identifies modulators of parkin recruitment to mitochondria. Journal of Biological Chemistry, 295(10), 3285–3300.
Moumné, L., Marie, A.-C., & Crouvezier, N. (2022). Oligonucleotide therapeutics: from discovery and development to patentability. Pharmaceutics, 14(2), 260.
Hu, B., Zhong, L., Weng, Y., Peng, L., Huang, Y., Zhao, Y., & Liang, X. J. (2020). Therapeutic siRNA: State of the art. Signal Transduction and Targeted Therapy, 5(1), 1–25.
Cavasotto, C. N., Lamas, M. S., & Maggini, J. (2021). Functional and druggability analysis of the SARS-CoV-2 proteome. European journal of pharmacology, 890, 173705.
Chaicumpa, W., Chaisri, U., Tapchaisri, P., Chongsa-Nguan, M., & Pongponratn, E. (1987). Oral vaccine against cholera prepared from Vibrio cholerae antigen(s). Southeast Asian Journal of Tropical Medicine and Public Health, 18(2), 142–148.
Koumakis, L. (2020). Deep learning models in genomics; are we there yet? Computational and Structural Biotechnology Journal, 18, 1466–1473.
Sato, K., Akiyama, M., & Sakakibara, Y. (2021). RNA secondary structure prediction using deep learning with thermodynamic integration. Nature communications, 12(1), 1–9.
Li, Y., Yang, X., Wang, N., Wang, H., Yin, B., Yang, X., & Jiang, W. (2020). GC usage of SARS-CoV-2 genes might adapt to the environment of human lung expressed genes. Molecular Genetics and Genomics, 295(6), 1537–1546.
Hagen, J. B. (2000). The origins of bioinformatics. Nature Reviews Genetics, 1(3), 231–236. https://doi.org/10.1038/35042090
Hogeweg, P. (2011). The roots of bioinformatics in theoretical biology. PLoS Computational Biology, 7(3), e1002021. https://doi.org/10.1371/journal.pcbi.1002021
Author information
Authors and Affiliations
Contributions
CC conceptualized the study, collected and analyzed the data, wrote the main manuscript. ARS did the validation and editing. MB and SC did formal analysis and developed the figures and tables. GA edited the manuscript. CC and SSL did the overall supervision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Research Involving Human Participants and/or Animals
Not applicable.
Informed Consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12033_2022_605_MOESM1_ESM.rar
Supplementary file1 (RAR 1691 kb)Fig. S1. RNA secondary structure model based on centroid of all nsp coding regions (nsp1–nsp16) components of the structure such as stem-loop, multi-branch loop etc. (A1) Secondary structure conformations of nsp1 coding gene and it components (A2) Secondary structure conformations of nsp2 coding gene and it components (A3) Secondary structure conformations of nsp3 coding gene and it components (A4) Secondary structure conformations of nsp4 coding gene (A5) Secondary structure conformations of nsp5 coding gene and it components (A6) Secondary structure conformations of nsp6 coding gene and it components (A7) Secondary structure conformations of nsp7 coding gene and it components (A8) Secondary structure conformations of nsp8 coding gene and it components (A9) Secondary structure conformations of nsp9 coding gene and it components (A10) Secondary structure conformations of nsp10 coding gene and it components (A11) Secondary structure conformations of nsp11 coding gene and it components (B1) Secondary structure conformations of nsp12 coding gene and it components (B2) Secondary structure conformations of nsp13 coding gene and it components (B3) Secondary structure conformations of nsp14 coding gene and it components (B4) Secondary structure conformations of nsp15 coding gene and it components (B5) Secondary structure conformations of nsp16 coding gene and it components
12033_2022_605_MOESM2_ESM.rar
Supplementary file2 (RAR 1503 kb)Fig. S2. RNA secondary structure model based on MFE of all nsp coding regions(nsp1–nsp16) (A1) Secondary structure conformations of nsp1 coding gene and it components (A2) Secondary structure conformations of nsp2 coding gene and it components (A3) Secondary structure conformations of nsp3 coding gene and it components (A4) Secondary structure conformations of nsp4 coding gene and it components (A5) Secondary structure conformations of nsp5 coding gene and it components (A6) Secondary structure conformations of nsp6 coding gene and it components (A7) Secondary structure conformations of nsp7 coding gene and it components (A8) Secondary structure conformations of nsp8 coding gene and it components (A9) Secondary structure conformations of nsp9 coding gene and it components (A10) Secondary structure conformations of nsp10 coding gene and it components (A11) Secondary structure conformations of nsp11 coding gene and it components (B1) Secondary structure conformations of nsp12 coding gene and it components (B2) Secondary structure conformations of nsp13 coding gene and it components (B3) Secondary structure conformations of nsp14 coding gene and it components (B4) Secondary structure conformations of nsp15 coding gene and it components (B5) Secondary structure conformations of nsp16 coding gene and it components
12033_2022_605_MOESM3_ESM.rar
Supplementary file3 (RAR 2522 kb)Fig. S3. Entropy and mountain plots based on the secondary structure of all nsp coding regions(nsp1–nsp16) (A1) Entropy and mountain plots of nsp1 coding gene (A2) Entropy and mountain plots of nsp2 coding gene (A3) Entropy and mountain plots of nsp3 coding gene (A4) Entropy and mountain plots of nsp4 coding gene (A5) Entropy and mountain plots of nsp5 coding gene (A6) Entropy and mountain plots of nsp6 coding gene (A7) Entropy and mountain plots of nsp7 coding gene (A8) Entropy and mountain plots of nsp8 coding gene (A9) Entropy and mountain plots of nsp9 coding gene (A10) Entropy and mountain plots of nsp10 coding gene (A11) Entropy and mountain plots of nsp11 coding gene (B1) Entropy and mountain plots of nsp12 coding gene (B2) Entropy and mountain plots of nsp13 coding gene (B3) Entropy and mountain plots of nsp14 coding gene (B4) Entropy and mountain plots of nsp15 coding gene (B5) Entropy and mountain plots of nsp16 coding gene
12033_2022_605_MOESM4_ESM.rar
Supplementary file4 (RAR 1363 kb)Fig. S4. A, T, G, C, A+T, and G+C content of the nsp coding genomic regions (A1) A, T, G, C, A+T, and G+C content of nsp1 coding gene (A2) A, T, G, C, A+T, and G+C content of nsp2 coding gene (A3) A, T, G, C, A+T, and G+C content of nsp3 coding gene (A4) A, T, G, C, A+T, and G+C content of nsp4 coding gene (A5) A, T, G, C, A+T, and G+C content of nsp5 coding gene (A6) A, T, G, C, A+T, and G+C content of nsp6 coding gene (A7) A, T, G, C, A+T, and G+C content of nsp7 coding gene (A8) A, T, G, C, A+T, and G+C content of nsp8 coding gene (A9) A, T, G, C, A+T, and G+C content of nsp9 coding gene (A10) A, T, G, C, A+T, and G+C content of nsp10 coding gene (A11) A, T, G, C, A+T, and G+C content of nsp11 coding gene (B1 A, T, G, C, A+T, and G+C content of nsp12 coding gene (B2) A, T, G, C, A+T, and G+C content of nsp13 coding gene (B3) A, T, G, C, A+T, and G+C content of nsp14 coding gene (B4) A, T, G, C, A+T, and G+C content of nsp15 coding gene (B5) A, T, G, C, A+T, and G+C content of nsp16 coding gene.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chakraborty, C., Bhattacharya, M., Sharma, A.R. et al. Structural Landscape of nsp Coding Genomic Regions of SARS-CoV-2-ssRNA Genome: A Structural Genomics Approach Toward Identification of Druggable Genome, Ligand-Binding Pockets, and Structure-Based Druggability. Mol Biotechnol 66, 641–662 (2024). https://doi.org/10.1007/s12033-022-00605-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00605-x