Abstract
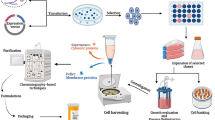
Survivin is one of the novel members of the apoptosis inhibitor protein family in humans. The main activity of the Survivin protein is to suppress caspases activity resulting in negative regulation of apoptosis. Survivin protein can be a potential target for the treatment of cancers between cancerous and normal cells. In the present research, the synthetic Survivin gene with PelB secretion signal peptide was cloned into a prokaryotic expression vector pET21a. The recombinant plasmid pET21a-PelB-Surv was expressed in Escherichia coli (E.coli) BL21, and the relative molecular mass of expressed protein was calculated 34,000 g/mol, approximately. The recombinant protein was purified through chromatography column and characterized by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Response surface methodology (RSM) was used to design 20 experiments for optimization of IPTG concentration, post-induction period, and cell density of induction (OD600). The optimum levels of the selected parameters were successfully determined to be 0.28 mM for IPTG concentration, 10 h for post-induction period, and 3.40768 for cell density (OD600). These findings resulted in 4.14-fold increases in the Survivin production rate of optimum expression conditions (93.6363 mg/ml).






Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
References
Sarela, A., Macadam, R., Farmery, S., Markham, A., & Guillou, P. (2000). Expression of the antiapoptosis gene, Survivin, predicts death from recurrent colorectal carcinoma. Gut, 46(5), 645–650.
Li, Y., Ma, X., Wu, X., Liu, X., & Liu, L. (2014). Prognostic significance of Survivin in breast cancer: Meta-analysis. The Breast Journal, 20(5), 514–524.
Goossens-Beumer, I., Zeestraten, E., Benard, A., Christen, T., Reimers, M., Keijzer, R., et al. (2014). Clinical prognostic value of combined analysis of Aldh1, Survivin, and EpCAM expression in colorectal cancer. British Journal of Cancer, 110(12), 2935–2944.
Chen, J., Li, T., Liu, Q., Jiao, H., Yang, W., Liu, X., et al. (2014). Clinical and prognostic significance of HIF-1α, PTEN, CD44v6, and Survivin for gastric cancer: A meta-analysis. PLoS ONE, 9(3), 91842.
Yie, S.-M., Lou, B., Ye, S.-R., He, X., Cao, M., Xie, K., et al. (2009). Clinical significance of detecting survivin-expressing circulating cancer cells in patients with non-small cell lung cancer. Lung Cancer, 63(2), 284–290.
Zaffaroni, N., & Daidone, M. G. (2002). Survivin expression and resistance to anticancer treatments: Perspectives for new therapeutic interventions. Drug Resistance Updates, 5(2), 65–72.
Cheung, C. H. A., Chen, H.-H., Kuo, C.-C., Chang, C.-Y., Coumar, M. S., Hsieh, H.-P., et al. (2009). Survivin counteracts the therapeutic effect of microtubule de-stabilizers by stabilizing tubulin polymers. Molecular Cancer, 8(1), 1–15.
Moriai, R., Tsuji, N., Moriai, M., Kobayashi, D., & Watanabe, N. (2009). Survivin plays as a resistant factor against tamoxifen-induced apoptosis in human breast cancer cells. Breast Cancer Research and Treatment, 117(2), 261–271.
Sun, X. P., Dong, X., Lin, L., Jiang, X., Wei, Z., Zhai, B., et al. (2014). Up-regulation of Survivin by AKT and hypoxia-inducible factor 1α contributes to cisplatin resistance in gastric cancer. The FEBS Journal, 281(1), 115–128.
McKenzie, J. A., Liu, T., Jung, J. Y., Jones, B. B., Ekiz, H. A., Welm, A. L., et al. (2013). Survivin promotion of melanoma metastasis requires upregulation of α 5 integrin. Carcinogenesis, 34(9), 2137–2144.
Zhang, M., Coen, J. J., Suzuki, Y., Siedow, M. R., Niemierko, A., Khor, L.-Y., et al. (2010). Survivin is a potential mediator of prostate cancer metastasis. International Journal of Radiation Oncology*Biology*Physics, 78(4), 1095–1103.
Di Stefano, A., Iovino, F., Lombardo, Y., Eterno, V., Höger, T., Dieli, F., et al. (2010). Survivin is regulated by interleukin-4 in colon cancer stem cells. Journal of Cellular Physiology, 225(2), 555–561.
Li, C., Yan, Y., Ji, W., Bao, L., Qian, H., Chen, L., et al. (2012). OCT4 positively regulates Survivin expression to promote cancer cell proliferation and leads to poor prognosis in esophageal squamous cell carcinoma. PLoS ONE, 7(11), e49693.
Yu, C.-J., Ou, J.-H., Wang, M.-L., Jialielihan, N., & Liu, Y.-H. (2015). Elevated Survivin mediated multidrug resistance and reduced apoptosis in breast cancer stem cells. Journal of B.U.ON, 20(5), 1287–1294.
Chantalat, L., Skoufias, D. A., Kleman, J.-P., Jung, B., Dideberg, O., & Margolis, R. L. (2000). Crystal structure of human Survivin reveals a bow tie–shaped dimer with two unusual α-helical extensions. Molecular Cell, 6(1), 183–189.
Jeyaprakash, A. A., Klein, U. R., Lindner, D., Ebert, J., Nigg, E. A., & Conti, E. (2007). Structure of a Survivin–Borealin–INCENP core complex reveals how chromosomal passengers travel together. Cell, 131(2), 271–285.
Vader, G., Kauw, J. J., Medema, R. H., & Lens, S. M. (2006). Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Reports, 7(1), 85–92.
Tsai, S.-L., Chang, Y.-C., Sarvagalla, S., Wang, S., Coumar, M. S., & Cheung, C. H. A. (2019). Cloning, expression, and purification of the recombinant pro-apoptotic dominant-negative survivin T34A–C84A protein in Escherichia coli. Protein Expression and Purification, 160, 73–83.
Baneyx, F., & Mujacic, M. (2004). Recombinant protein folding and misfolding in Escherichia coli. Nature Biotechnology, 22(11), 1399–1408.
Soares, C. R., Gomide, F. I., Ueda, E. K., & Bartolini, P. (2003). Periplasmic expression of human growth hormone via plasmid vectors containing the λPL promoter: Use of HPLC for product quantification. Protein Engineering, 16(12), 1131–1138.
Arora, D., & Khanna, N. (1996). Method for increasing the yield of properly folded recombinant human gamma interferon from inclusion bodies. Journal of Biotechnology, 52(2), 127–133.
Rai, M., & Padh, H. (2001). Expression systems for production of heterologous proteins. Current Science, 80(9), 1121–1128.
Lim, H. K., & Jung, K. H. (1998). Improvement of heterologous protein productivity by controlling postinduction specific growth rate in recombinant Escherichia coli under control of the PL promoter. Biotechnology Progress, 14(4), 548–553.
Arthur, P. M., Duckworth, B., & Seidman, M. (1990). High level expression of interleukin-1β in a recombinant Escherichia coli strain for use in a controlled bioreactor. Journal of Biotechnology, 13(1), 29–46.
Shin, C. S., Hong, M. S., Bae, C. S., & Lee, J. (1997). Enhanced production of human mini-proinsulin in fed-batch cultures at high cell density of Escherichia coli BL21 (DE3)[pET-3aT2M2]. Biotechnology Progress, 13(3), 249–257.
Yoon, S. K., Kang, W. K., & Park, T. H. (1994). Fed-batch operation of recombinant Escherichia coli containing trp promoter with controlled specific growth rate. Biotechnology and Bioengineering, 43(10), 995–999.
Narum, D. L., Kumar, S., Rogers, W. O., Fuhrmann, S. R., Liang, H., Oakley, M., et al. (2001). Codon optimization of gene fragments encoding plasmodium falciparum merzoite proteins enhances DNA vaccine protein expression and immunogenicity in mice. Infection and Immunity, 69(12), 7250–7253.
Maldonado, L., Hernández, V., Rivero, E., de la Rosa, A. B., Flores, J. L. F., Acevedo, L. G. O., & De León Rodríguez, A. (2007). Optimization of culture conditions for a synthetic gene expression in Escherichia coli using response surface methodology: The case of human interferon beta. Biomolecular Engineering, 24, 217–222.
Chen, H.-C., Hwang, C.-F., & Mou, D.-G. (1992). High-density Escherichia coli cultivation process for hyperexpression of recombinant porcine growth hormone. Enzyme and Microbial Technology, 14(4), 321–326.
Nikerel, I. E., Öner, E., Kirdar, B., & Yildirim, R. (2006). Optimization of medium composition for biomass production of recombinant Escherichia coli cells using response surface methodology. Biochemical Engineering Journal, 32(1), 1–6.
Baskar, G., Dharmendira Kumar, M., Anand Prabu, A., Renganathan, S., & Yoo, C. (2009). Optimization of carbon and nitrogen sources for L-asparaginase production by Enterobacter aerogenes using response surface methodology. Chemical and Biochemical Engineering Quarterly, 23(3), 393–397.
Transformation B. The Heat Shock Method. JoVE Science Education Database Basic Methods in Cellular and Molecular Biology. 2015.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227(5259), 680–685.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72(1–2), 248–254.
Bokarewa, M., Lindblad, S., Bokarew, D., & Tarkowski, A. (2005). Balance between Survivin, a key member of the apoptosis inhibitor family, and its specific antibodies determines erosivity in rheumatoid arthritis. Arthritis Research & Therapy, 7(2), 349.
Zhang, W., Gao, Y., Li, F., Tong, X., Ren, Y., Han, X., et al. (2015). YAP promotes malignant progression of Lkb1-deficient lung adenocarcinoma through downstream regulation of Survivin. Cancer Research, 75(21), 4450–4457.
Sarela, A., Verbeke, C., Ramsdale, J., Davies, C., Markham, A., & Guillou, P. (2002). Expression of Survivin, a novel inhibitor of apoptosis and cell cycle regulatory protein, in pancreatic adenocarcinoma. British Journal of Cancer, 86(6), 886–892.
Zhang, M., Latham, D. E., Delaney, M. A., & Chakravarti, A. (2005). Survivin mediates resistance to antiandrogen therapy in prostate cancer. Oncogene, 24(15), 2474–2482.
Falleni, M., Pellegrini, C., Marchetti, A., Oprandi, B., Buttitta, F., Barassi, F., et al. (2003). Survivin gene expression in early-stage non-small cell lung cancer. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland., 200(5), 620–626.
Brierley, R. A., Davis, G. R., Holtz, G. C. (1994). Production of insulin-like growth factor-1 in methylotrophic yeast cells. Google Patents.
Gopal, G. J., & Kumar, A. (2013). Strategies for the production of recombinant protein in Escherichia coli. The Protein Journal, 32(6), 419–425.
Gray, G. L., Baldridge, J. S., McKeown, K. S., Heyneker, H. L., & Chang, C. N. (1985). Periplasmic production of correctly processed human growth hormone in Escherichia coli: Natural and bacterial signal sequences are interchangeable. Gene, 39(2–3), 247–254.
Meng, L., Xi, J., & Yeung, M. (2016). Degradation of extracellular polymeric substances (EPS) extracted from activated sludge by low-concentration ozonation. Chemosphere, 147, 248–255.
Shokri, A., Sandén, A., & Larsson, G. (2003). Cell and process design for targeting of recombinant protein into the culture medium of Escherichia coli. Applied Microbiology and Biotechnology, 60(6), 654–664.
Tong, Y., & Lighthart, B. (2000). The annual bacterial particle concentration and size distribution in the ambient atmosphere in a rural area of the Willamette Valley, Oregon. Aerosol Science & Technology, 32(5), 393–403.
Choi, J., & Lee, S. (2004). Secretory and extracellular production of recombinant proteins using Escherichia coli. Applied Microbiology and Biotechnology, 64(5), 625–635.
Galloway, C. A., Sowden, M. P., & Smith, H. C. (2003). Increasing the yield of soluble recombinant protein expressed in E. coli by induction during late log phase. Biotechniques, 34(3), 524–530.
Chae, H. J., DeLisa, M. P., Cha, H. J., Weigand, W. A., Rao, G., & Bentley, W. E. (2000). Framework for online optimization of recombinant protein expression in high-cell-density Escherichia coli cultures using GFP-fusion monitoring. Biotechnology and Bioengineering., 69(3), 275–285.
De Marco, A. (2009). Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microbial Cell Factories, 8(1), 1–18.
Pratap, J., Rajamohan, G., & Dikshit, K. L. (2000). Characteristics of glycosylated streptokinase secreted from Pichia pastoris: Enhanced resistance of SK to proteolysis by glycosylation. Applied Microbiology and Biotechnology, 53(4), 469–475.
Aneja, R., Datt, M., Singh, B., Kumar, S., & Sahni, G. (2009). Identification of a new exosite involved in catalytic turnover by the streptokinase-plasmin activator complex during human plasminogen activation. Journal of Biological Chemistry, 284(47), 32642–32650.
Chen, R. (2012). Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnology Advances, 30(5), 1102–1107.
Francis, D. M., & Page, R. (2010). Strategies to optimize protein expression in E. coli. Current Protocols in Protein Science. https://doi.org/10.1002/0471140864.ps0524s61
Xie, L., Hall, D., Eiteman, M., & Altman, E. (2003). Optimization of recombinant aminolevulinate synthase production in Escherichia coli using factorial design. Applied Microbiology and Biotechnology., 63(3), 267–273.
Larentis, A. L., Argondizzo, A. P. C., dos Santos, E. G., Jessouron, E., Galler, R., & Medeiros, M. A. (2011). Cloning and optimization of induction conditions for mature PsaA (pneumococcal surface adhesin A) expression in Escherichia coli and recombinant protein stability during long-term storage. Protein Expression and Purification, 78(1), 38–47.
Zamani, M., Berenjian, A., Hemmati, S., Nezafat, N., Ghoshoon, M. B., Dabbagh, F., et al. (2015). Cloning, expression, and purification of a synthetic human growth hormone in Escherichia coli using response surface methodology. Molecular biotechnology, 57(3), 241–250.
Funding
This study was funded by the Agricultural and Natural Resources and Education Research Center (AREEO), Kerman, Iran.
Author information
Authors and Affiliations
Contributions
LYG participated in the conceptualization, study design, planning, methodology, and execution of the study. The data gathered were reviewed and edited by NR. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rostami, N., Goharrizi, L.Y. Cloning, Expression, and Purification of the Human Synthetic Survivin Protein in Escherichia coli Using Response Surface Methodology (RSM). Mol Biotechnol 65, 326–336 (2023). https://doi.org/10.1007/s12033-021-00399-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-021-00399-4