Abstract
Human papillomaviruses (HPV)-16 and 18 are the most prevalent types associated with cervical cancer. HPV L1 and L2 capsid proteins and E7 oncoprotein play crucial roles in HPV-related diseases. Hence, these proteins were proposed as target antigens for preventive and therapeutic vaccines. In this study, two multiepitope DNA-based HPV vaccine candidates were designed using in silico analysis including the immunogenic and conserved epitopes of HPV16/18 L1, L2 and E7 proteins (the L1-L2-E7 fusion DNA), and of heat shock protein 70 (HSP70) linked to the L1-L2-E7 DNA construct (the HSP70-L1-L2-E7 fusion DNA). Next, the expression of the L1-L2-E7 and HSP70-L1-L2-E7 multiepitope DNA constructs was evaluated in a mammalian cell line. Finally, immunological responses and antitumor effects of the DNA constructs were investigated in C57BL/6 mice. Our data indicated high expression rates of the designed multiepitope L1-L2-E7 DNA (~ 56.16%) and HSP70-L1-L2-E7 DNA (~ 80.45%) constructs in vitro. The linkage of HSP70 epitopes to the L1-L2-E7 DNA construct significantly increased the gene expression. Moreover, the HSP70-L1-L2-E7 DNA construct could significantly increase immune responses toward Th1 response and CTL activity, and induce stronger antitumor effects in mouse model. Thus, the designed HSP70-L1-L2-E7 DNA construct represents promising results for development of HPV DNA vaccine candidates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infectious agents are responsible for 20–25% of all cancer cases in the world [1]. Among them, about 15% of human cancers were related to viral infections [2, 3]. For example, human papillomavirus (HPV) caused ~ 30% of all infectious agents-related cancers, and was associated with more than 95% of cervical carcinomas [4,5,6]. The genome of HPV is divided into two main regions encoding early (E) and late (L) proteins. The early proteins regulate the viral DNA replication in the basal layer of epithelial cells, and the late proteins make viral capsid [7,8,9]. Among early proteins, E6 and E7 oncoproteins are required for the initiation of HPV-associated malignancies and are expressed in transformed cells. Therefore, HPV E6 and E7 oncoproteins serve as ideal targets for therapeutic HPV vaccines [10].
Among more than 200 recognized HPV genotypes, two high-risk HPV types such as HPV16 and HPV18 have been recognized as the most prevalent types related to nearly 70% of cervical cancers and precancerous cervical lesions [11, 12]. Hence, considerable efforts have been made to control HPV-induced diseases using prophylactic or therapeutic approaches [13, 14]. Up to now, three prophylactic HPV vaccines were FDA-approved based on viral like particles (VLPs) composed of L1 protein including Cervarix (bivalent HPV16/18 vaccine), Gardasil (quadrivalent HPV16/18/6/11 vaccine), and Gardasil-9 (nonavalent HPV16/18/31/33/45/52/58/30/40 vaccine) [15]. Although, these vaccines were effective only in subjects who were not previously exposed to HPV, but none of them did not show therapeutic effects on the established HPV infection and associated cancers. Furthermore, their high cost is a major problem in low-income countries [16, 17]. Therefore, development of strong and potent therapeutic vaccine is vital, as well.
Various types of therapeutic vaccines including live bacterial/viral vectors-, RNA/DNA-, protein/peptide- and cell-based vaccines have been tested to treat the HPV-associated diseases [18,19,20]. For example, some vaccines have passed clinical trials including SGN-00101 protein vaccine (composed of heat shock protein fused to HPV16 E7), ZYC-101a DNA vaccine (composed of HPV16/18 E6 and E7), HPV16 L1-E7 chimeric VLP, TA-HPV (composed of recombinant vaccinia virus expressing E6 and E7), TA-CIN protein vaccine (composed of fusion E6-E7-L2 protein), and PC10VAC01 (composed of HPV16 E7 + adenylate cyclase) [21]. However, each vaccination approach indicated some benefits and limitations [18]. Thus, it is crucial to find the potent and safe strategies in developing therapeutic vaccine and enhancing their immunogenicity [22, 23].
Some studies reported the benefits of heat shock proteins (HSPs) as an adjuvant to increase the antitumor potency of vaccines. HSPs effectively stimulate both innate and adaptive immunity [24, 25]. Among various types of HSPs, HSP70 is a promising molecule because of its adjuvant activity to increase the antigen-specific immunity [24,25,26,27,28,29]. As known, immunoinformatics tools may assist scientists to predict high immunogenic and conserved epitopes, which induce B- or T-cell responses against HPV infection [20, 30,31,32,33,34,35]. Moreover, linkage of antigens to HSPs provided a promising strategy to increase the efficiency of vaccine candidates [29, 36].
In current study, in silico approaches were used to design the multiepitope L1-L2-E7, and HSP70-L1-L2-E7 constructs as novel and potent vaccine candidates. The E7 protein was used alone or combined with other HPV proteins especially E6 protein for development of therapeutic clinical trials [21]. Herein, we used E7 protein along with L1 and L2 proteins for design of vaccine constructs. For in vitro assay, the expression of both multiepitope DNA constructs was studied in a mammalian cell line. Finally, the immunological and antitumor effects of both DNA constructs were investigated in C57BL/6 mice.
Materials and Methods
Materials
The restriction enzymes and DNA or protein ladder were purchased from Fermentas Company. DNA extraction kits were prepared from Qiagen Company. The cell culture medium, serum, antibiotic were purchased from Gibco or Biosera Company. The cell lines were provided from the cell bank at Pasteur Institute of Iran. The conjugated antibodies, cytokine and granzyme B assay kits were purchased from Sigma, Mabtech and eBioscience Company, respectively.
Immunoinformatics Analyses
Protein Sequences
The reference protein sequence of heat shock 70 kDa protein 1A (HspA1A; NP_005336.3) was obtained from the National Center for Biotechnology Information Database (NCBI) (http://www.ncbi.nlm.nih.gov/). The reference HPV16/18 L1, L2, E7 protein sequences were previously determined from NCBI and UniProtKB/Swiss-Prot database, and used for bioinformatics analyses by our group [20, 33, 34].
Plan of the Study
In order to determine the potential CD8+ and CD4+ T-cell epitopes, a two-step plan was designed (Figs. 1 and 2). Briefly, the first step included (a) Epitope prediction bound to MHC-I or MHC-II, (b) TAP transport/proteasomal cleavage analysis, (c) Immunogenicity, allergenicity and toxicity analyses, (d) Prediction of cytokine induction, (e) Population coverage, and (f) Protein-peptide docking analyses (Fig. 1). This step was previously performed for epitope prediction of HPV 16 & 18 L1, L2 and E7 proteins [20, 33, 34]. Herein, we determined the immunogenic and conserved epitopes of HSP70. In the second step, two multiepitope peptide constructs were designed and examined for physicochemical characteristics, protein solubility, B-cell epitope prediction, secondary and 3D structure modeling, refinement of 3D structure, validation of the refined 3D structure, and molecular docking between toll-like receptors and novel constructs (Fig. 2).
HLA Allele Frequency
The frequency of HLA supertypes and alleles was determined from allele frequency net database (AFND) (http://www.allelefrequencies.net/).
T-Cell Epitope Prediction
Determination of T-cell immunodominant epitopes was known as the most critical step for design of a multiepitope-based vaccine candidate using immune-informatics tools [37, 38]. Herein, the design of the multiepitope construct harboring HLA-class I restricted cytotoxic T lymphocyte (CTL) epitopes and also HLA-class II restricted helper T lymphocyte (HTL) epitopes was performed as follows.
MHC-I Binding Prediction
Binding of MHC class I molecules to epitopes is followed by antigen presentation to CTLs. Herein, IEDB MHC-I prediction (http://tools.iedb.org/mhci/) and NetMHCpan4.1 (http://www.cbs.dtu.dk/services/NetMHCpan/) web servers were applied to predict the binding of peptides (8–11 residues) to MHC class I molecules (default thresholds: 0.5% and 2% for strong and weak binders, respectively) [39]. For prediction of T-cell epitopes of HSP70 bound to human and mouse MHC alleles, IEDB MHC-I prediction tool (IEDB recommended method) was applied, as well. Predictions were performed against H2-Db, H2-Dd, H2-Kb, H2-Kd, H2-Kk, H2-Ld, H2-Qa1 and H2-Qa2 MHC-I mouse alleles.
MHC-II Binding Prediction
Binding of MHC class II molecules to epitopes is critical for the HTL activation [40]. IEDB MHC-II binding prediction tool (http://tools.iedb.org/mhcii/; IEDB recommended method) and NetMHCIIpan 4.0 web server (http://www.cbs.dtu.dk/services/NetMHCIIpan; default thresholds: 2% and 10% for strong and weak binders, respectively) were applied to predict the binding of peptides to MHC-II molecules.
TAP Transport/Proteasomal Cleavage Analysis
The best ranked peptides obtained from IEDB, NetMHCpan4.1, and NetMHCIIpan databases were employed in transporter associated with antigen presentation (TAP), and proteasomal cleavage analyses. Herein, IEDB tool (http://tools.iedb.org/processing/) and NetCTL1.2 server (http://www.cbs.dtu.dk/services/NetCTL/) were utilized to predict antigen processing in MHC-I presentation pathway. The higher TAP score shows the higher transport rate [41, 42].
MHC-I Immunogenicity Prediction
The IEDB web server (http://tools.iedb.org/immunogenicity/) [43] was applied to determine the MHC-I immunogenicity of the predicted epitopes.
Allergenicity and Toxicity Analyses
An efficient, potent and safe vaccine candidate is not allergic. Herein, the allergenicity and toxicity of the selected epitopes were analyzed by AlgPred server (https://webs.iiitd.edu.in/raghava/algpred/) [44] and ToxinPred web server (https://webs.iiitd.edu.in/raghava/toxinpred/) [45], respectively.
Cytokine Induction-Based Prediction
The chosen epitopes were submitted to evaluate whether they can stimulate Interleukine-10 (IL-10), Interleukine-4 (IL-4), and IFN-gamma using (https://webs.iiitd.edu.in/raghava/il10pred/), (https://webs.iiitd.edu.in/raghava/il4pred/), and (https://webs.iiitd.edu.in/raghava/ifnepitope/) web servers, respectively [46,47,48].
Population Coverage
Selection of the multiple peptides with different HLA binding specificities will increase the population coverage targeted by peptide-based vaccines [49]. Population coverage for each epitope and its binding to HLA alleles in various geographic areas were analyzed by IEDB population coverage tool (http://tools.iedb.org/population/).
MHC-Peptide Docking
The peptide-protein (e.g., peptide-MHC) interaction is the major goal of computational docking [50]. For prediction of 3D MHC-peptide complex structures, both human and mouse MHC alleles were considered using GalaxyPepDock server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=PEPDOCK) based on interaction similarity and energy optimization [51]. The RCSB PDB server (https://www.rcsb.org/) was used to access the available PDB files of HLA alleles.
Construct Design
For design of the multiepitope peptide constructs, immunoinformatics tools were utilized to select novel immunodominant T-cell epitopes. The selected T-cell epitopes of HPV16/18 L1, L2 & E7 proteins [20, 33, 34] were used to design a novel construct fused by AAY linker (the L1-L2-E7 fusion multiepitope peptide construct; Fig. 3A). In addition, the immunodominant T-cell epitopes of HSP70 were linked to the L1-L2-E7 construct for design of the second multiepitope peptide construct (the HSP70-L1-L2-E7 multiepitope construct; Fig. 3B). The HSP70 epitopes as an immune stimulating agent can enhance the immunogenicity of vaccine construct.
B-Cell Epitope Prediction
An effective multiepitope-based vaccine candidate should possess a favorite secondary structure for induction of the peptide-specific humoral response, as well [52]. Thus, both constructs were applied to predict B-cell epitopes using IEDB Bepipred Linear Epitope Prediction (http://tools.iedb.org/bcell/) [53].
Physicochemical Properties
Various physicochemical properties and the solubility of two constructs were predicted by ProtParam server tools [54] and protein-sol (https://protein-sol.manchester.ac.uk/) web server, respectively.
Modeling the Secondary Structure
The secondary structures of both constructs were predicted by Predict Secondary Structure (PSIPRED) (http://bioinf.cs.ucl.ac.uk/psipred/) and RaptorX (http://raptorx.uchicago.edu/StructurePropertyPred/predict/) servers. RaptorX Property web server predicts the secondary structure using DeepCNF machine learning model [55].
Modeling, Refinement and Validation of 3D Structures
The tertiary structures of both constructs were predicted by Iterative Threading ASSEmbly Refinement (I-TASSER) server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) [56]. Then, the refinement of Top 3D structure model determined from I-TASSER was done using GalaxyRefine Server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE) based on molecular dynamics simulation [57, 58]. Finally, validation and selection of the best models of a refined structure were analyzed by ERRAT server (https://servicesn.mbi.ucla.edu/ERRAT/) [59].
Molecular Docking Between the Multiepitope Constructs and Toll-Like Receptors
Molecular docking between the multiepitope peptide constructs and various toll-like receptors (TLRs) was done to predict the possible binding orientation of the multiepitope constructs using ClusPro 2.0 (https://cluspro.bu.edu) [60]. For protein–protein docking, the final refined tertiary structures of the designed vaccine constructs were submitted as ligands [61, 62]. Additionally, the PDB files of TLRs were received from RCSB at https://www.rcsb.org.
In Vitro Analysis
Preparation of the HSP70-L1-L2-E7 DNA Fusion Construct
At first, a general multiepitope peptide construct was designed as shown in Fig. 4. Then, the nucleotide sequence of HSP70-L1-L2-E7 was retrieved by amino acid reverse translation tool (http://www.bioinformatics.org/sms2/rev_trans.html), and the restriction enzyme sites were determined for cloning process. Next, the HSP70-L1-L2-E7 DNA construct was synthesized in pUC57 cloning vector by Bio Magic Gene Company. After that, the L1-L2-E7 and HSP70-L1-L2-E7 genes were subcloned into the BglII/HindIII and XhoI/HindIII cloning sites of the pEGFP-N1 eukaryotic expression vector, respectively for in vitro studies. In general, the cloning step included digestion of vector and the target gene with the restriction enzymes, gel extraction of the linearized vector and the insert, ligation of the insert and vector by T4 DNA ligase (Fermentas), transformation of the ligation product in E. coli DH5α strain, and extraction and confirmation of the recombinant plasmid. Moreover, the L1-L2-E7 and HSP70-L1-L2-E7 genes were subcloned into the EcoRI/HindIII and BamHI/HindIII cloning sites of the pcDNA3.1 (-) eukaryotic expression vector (cytomegalovirus “CMV” promoter), respectively for in vivo studies. Finally, the recombinant endotoxin-free plasmids (i.e., pEGFP-L1-L2-E7, pEGFP-HSP70-L1-L2-E7, pcDNA-L1-L2-E7, and pcDNA-HSP70-L1-L2-E7) were prepared using a Maxi-Kit DNA extraction (Qiagen). Their concentration and purity were revealed by NanoDrop spectrophotometry.
In Vitro Expression of the L1-L2-E7 and HSP70-L1-L2-E7 DNA Constructs in Mammalian Cells
Human embryonic kidney 293T (HEK-293T) cells were cultured in RPMI 1640 medium (Gibco), supplemented with 10% FBS (Fetal Bovine Serum; Biosera, France), 1% Penstrep (Sigma, Germany) at 37 °C and 5% CO2 in a 24-well plate. When the confluency of the cells reached approximately 80%, the cells were transfected with the TurboFect-plasmid DNA complexes. Herein, 2 μL of TurboFect (Termo Scientifc) and 1 μg of pEGFP-L1-L2-E7, pEGFP-HSP70-L1-L2-E7 or pEGFP-N1 (as a positive control) were mixed and incubated for 20 min at room temperature to form the TurboFect-plasmid DNA complexes. The expression of DNA constructs was analyzed at 48 h after transfection using flow cytometry, fluorescent microscopy, and Western blotting. In Western blotting, the anti-GFP polyclonal antibody (1:5000 v/v; Abcam), and 3,3′-diaminobenzidine (DAB, Sigma) substrate were used to recognize the expressed proteins, and detect the immunoreactive protein bands, respectively.
In Vivo Studies
The Synthesis of Peptide Constructs
For immunological assay, two multiepitope peptide constructs (L1-L2-E7, and HSP70-L1-L2-E7, Fig. 3) were synthesized by BioMatik Company.
Immunization of Mice
Four groups of eight female C57BL/6 mice (maintained at Pasteur Institute of Iran under specific pathogen-free conditions) were injected on days 0, 14, and 28 with the plasmid DNA (pcDNA-L1-L2-E7 or pcDNA-HSP70-L1-L2-E7: G1 or G2; 50 μg) subcutaneously at the right footpad (Table 1). The control groups (G3 and G4) were injected with pcDNA3.1 and PBS, respectively. The animal experimental procedures were approved by Animal Care and Use Committee of Islamic Azad University-Science and Research Branch, and performed according to the Animal Experimentation Regulations of Islamic Azad University for scientific purposes (Ethics code: IR.IAU.SRB.REC.1398.208; Approval date: 2020-02-22).
Monitoring Tumor Growth
For in vivo preventive test, vaccinated mice with different regimens were subcutaneously challenged in the right flank with 1 × 105 HPV16-expressing C3 tumor cells, 3 weeks after the third injection. Tumor cell line C3 was generated by transfection of mouse embryonic cells with the complete HPV genome and maintained as previously described [63]. Tumor growth and the percentage of tumor-free mice were assessed twice a week by palpation for 65 days after C3 challenge. At each time point, tumor volume was calculated using the formula: V = (a2b)/2 (a: width, b: length).
Antibody Assay
The mice sera were prepared from each group 3 weeks after the third injection. The levels of goat anti-mouse immunoglobulin G1 (IgG1), IgG2a and total IgG antibodies (1:10,000 v/v, Sigma) were assessed in the pooled sera of each group (1:100 v/v) using indirect ELISA. The coated antigens were the HSP70-L1-L2-E7, and L1-L2-E7 synthetic peptides (5 μg/mL).
Cytokine Assay
Three weeks after the last injection, the red blood cell-depleted pooled splenocytes (2 × 106 cells/mL) of three mice from each group were cultured in 48-well plates for 72 h in the presence of 5 μg/mL of the L1-L2-E7 or HSP70-L1-L2-E7 peptides, negative control (RPMI 5%), and positive control (Concanavalin A: 5 μg/mL). The secretion of IFN-γ, IL-4 and IL-10 was assessed in the supernatants using the sandwich ELISA kit (Mabtech Swedish Biotech Co.). The results were shown as mean ± SD for each group.
Granzyme B Assay (In Vitro CTL Activity)
The P815 target cells (T: 2 × 104 cells/well) were incubated with the L1-L2-E7 or HSP70-L1-L2-E7 peptides (~ 30 μg/mL) for 24 h. Then, the effector cells (E: the red blood cell-depleted pooled splenocytes) were added to the target cells at T/E ratio of 1/100, and incubated for 6 h. Finally, the concentration of Granzyme B (GrB) was assessed in the supernatants using ELISA (eBioscience kit).
Therapeutic Effects
For therapeutic tests of the established C3 tumors, four groups of five female C57BL/6 mice (similar to Table 1) were considered. Briefly, five mice in each group were subcutaneously injected with 1 × 105 C3 tumor cells, and then 1 week after tumor challenge, mice received various regimens (Table 1, the dose of DNA constructs: 50 μg) three times with a 2-week interval. Finally, tumor growth was detected two times a week for 65 days.
Statistical Analysis
Statistical analysis was done by Prism 7.0 software (GraphPad) using one-way ANOVA and Student’s t-test. The percentage of tumor-free mice (or survival rate) was determined by the log-rank test. The p-value < 0.05 was statistically considered significant. The experiments were independently performed twice.
Results
In Silico Studies
T-Cell Epitope Prediction
T-cell epitope prediction for the HPV16/18 L1, L2 and E7 proteins was previously done by our group [20, 33, 34]. In this study, the best epitopes were used to design of a fusion construct (L1-L2-E7) for in silico, in vitro and in vivo analyses. Moreover, HSP70 (HspA1A: NP_005336.3) protein sequence was analyzed to identify the most putative immunogenic and conserved regions. The peptides derived from HSP70 were selected based on the strongest binding affinity to human and mouse MHC class I and II alleles. The predicted MHC-I and MHC-II epitopes are listed in Tables 2 and 3.
TAP Transport/Proteasomal Cleavage
TAP transport efficiency and proteasomal cleavage scores were previously determined for L1, L2 and E7 proteins [20, 33, 34]. Herein, HSP70 113FYPEEISSMVLTKM126 and 285SLFEGIDFYTSITR298 epitopes had the highest epitope identification scores as shown in Table 4.
Immunogenicity, Toxicity and Allergenicity Analyses
Immunogenicity, toxicity and allergenicity for the L1, L2 and E7 epitopes of HPV types 16 and 18 were analyzed in our previous studies [20, 33, 34]. Herein, these analyses were performed for the selected Hsp70 epitopes as shown in Table 5. As observed, these peptides were non-allergen and non-toxic.
Cytokine Analysis
The MHC class II binding epitopes of HPV16/18 L1, L2 and E7 proteins as well as HSP70 were analyzed for the possible induction of IFN-γ, IL-10 and IL-4 cytokines. The higher rates showed more potent epitopes for inducing cytokines as shown in Table 6. For instance, all candidate epitopes of HPV16/18 L1, L2 and E7 proteins and HSP70 induced IFN-γ cytokine.
Population Coverage Analysis
In our previous study, population coverage was calculated for the selected HPV16/18 L1, L2 and E7 epitopes [20, 33, 34]. Herein, the highest population coverage was determined for HSP70 epitopes in different area. The rates were 88.39% and 81.19% for CTL epitopes of HSP70285–298 and HSP70113–126, and 21.86% and 45.07% for HTL epitopes of HSP168–182 and HSP70389–403, respectively in the world’s population (Table 7).
Molecular Docking
The human and mouse MHC alleles used for molecular docking against the selected HSP70 peptides are shown in Table 8. Moreover, the interaction similarity scores of human and mouse MHC-I/MHC-II alleles were determined for HSP70 epitopes as shown in Tables 9, 10, 11, 12. Figures 5 and 6 are successful examples of molecular MHC-peptide docking between the selected peptides and human/mouse MHC-I and MHC-II alleles, respectively. These analyses were performed for HPV16/18 L1, L2 and E7 epitopes in our previous study [20, 33, 34].
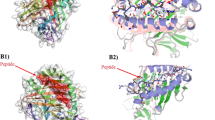
Peptide-MHC docking between the selected epitopes of HSP70 and human MHC alleles: A The successful docking between 113FYPEEISSMVLTKM126 and HLA-A2402 with interaction score of 255.0; B The successful docking between 285SLFEGIDFYTSITR298 and HLA-A0201 with interaction score of 258.0; C The successful docking between 168NVLRIINEPTAAAIA182 and DRB1-0101 with interaction score of 145.0; D The successful docking between 389QDLLLLDVAPLSLGL403 and DRB1-0101with interaction score of 158.0
Peptide-MHC docking between the selected epitopes of HSP70 and mouse MHC alleles: A The successful docking between 113FYPEEISSMVLTKM126 and H-2-Db with interaction score of 344.0; B The successful docking between 285SLFEGIDFYTSITR298 and H-2-Db with interaction score of 314.0; C The successful docking between 168NVLRIINEPTAAAIA182 and H-2-IAb with interaction score of 107.0; D The successful docking between 389QDLLLLDVAPLSLGL403 and H-2-IAb with interaction score of 107.0
Construct Design
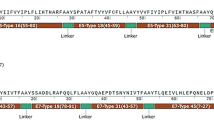
The selected CTL epitopes include HPV16 L1 (12YLPPVPVSKV21), HPV16 L2 (11KRASATQLYK20), HPV16 E7 (216GQAEPDRAHYNIVTF230), HPV18 L1 (461DQYPLGRKFLV471), HPV18 L2 (11KRASVTDLYK20), HPV18 E7 (78SSADDLRAFQQLFL91), HSP70 (113FYPEEISSMVLTKM126), and HSP70 (285SLFEGIDFYTSITR298). The selected HTL epitopes contain HPV16 L1 (416DTYRYVQSQAITCQK430), HPV16 L2 (281PDFLDIVALHRPALTSR297), HPV18 L1 (8DNTVYLPPPSVARVV22), HPV18 L2 (274SDFMDIIRLHRPALTSR290), HSP70 (168NVLRIINEPTAAAIA182), and (389QDLLLLDVAPLSLGL403). These top-ranked epitopes were fused by AAY linker for design of the L1-L2-E7 and HSP70-L1-L2-E7 multiepitope peptide constructs (Fig. 3).
B-Cell Epitope Prediction
The sequences and position of the predicted B-cell epitopes are shown in Fig. 7 and Table 13. These regions had the ability to induce antibody.
Physicochemical Characteristics
Various physicochemical properties of the L1-L2-E7 and HSP70-L1-L2-E7 constructs were determined by ProtParam tool as shown in Table 14. Our results indicated that the L1-L2-E7 and HSP70-L1-L2-E7 multiepitope peptides had molecular weight (MW) of about 19.5 kDa and 28.5 kDa, respectively. Moreover, the L1-L2-E7 and HSP70-L1-L2-E7 multiepitope peptides were unstable with the instability index (II) of 41.53 and 43.63 (i.e., a value more than 40 means that the protein is not stable [64]), and also were less soluble with the probability of 0.376 and 0.301, respectively.
Secondary Structure Modeling
The secondary structure prediction of the L1-L2-E7 and HSP70-L1-L2-E7 multiepitope peptide constructs was done by PSIPRED and RaptorX servers as shown in Fig. 8. The secondary structure for the L1-L2-E7 construct was composed of 42% α-helix, 6% β-sheet, 51% coil, and 5% disordered. The secondary structure for the HSP70-L1-L2-E7 construct was composed of 43% α-helix, 17% β-sheet, 38% coil, and 3% disordered. Linkage of HSP70 epitopes changed the rates of β-sheet and coil secondary structures.
3D Structure Prediction, Refinement and Validation
The assurance of each 3D model predicted by I-TASSER was calculated using C-score that is commonly in the range of − 5 to 2. A higher value of C-score shows a higher confidence for a model. The C-scores of the best model for the L1-L2-E7 and HSP70-L1-L2-E7 multiepitope peptides were − 3.66 and − 2.72, respectively. Figure 9 indicates the tertiary structures of the predicted L1-L2-E7 and HSP70-L1-L2-E7 constructs. The top model of each construct was refined by GalaxyRefine server. Then, the top refined model was validated by ERRAT server. Our results showed that the quality of 3D structure was improved after refinement process as shown in Table 15 and Fig. 10.
The 3D structures of the L1-L2-E7 (A) and HSP70-L1-L2-E7 (B) constructs predicted by I-TASSAR server: A The L1-L2-E7 construct with C-score of -3.66 has the highest score among the predicted structures; B The HSP70-L1-L2-E7 construct with C-score of − 2.72 has the highest score among the predicted structures
Molecular Docking Between the Multiepitope Peptides and TLRs
Molecular docking between the multiepitope constructs and TLRs was done using ClusPro 2.0. After prediction, we selected the models which properly occupied the receptor and had the lowest energy scores. The lowest energy level obtained for TLR-2-L1-L2-E7, TLR-3-L1-L2-E7, TLR-4-L1-L2-E7, TLR-5-L1-L2-E7, TLR-8-L1-L2-E7 and TLR-9-L1-L2-E7 constructs was − 839.0, − 957.8, − 908.0, − 1210.0, − 929.0, and − 1034.0, respectively as shown in Fig. 11. The lowest energy level achieved for TLR-2-HSP70-L1-L2-E7, TLR-3-HSP70-L1-L2-E7, TLR-4-HSP70-L1-L2-E7, TLR-5-HSP70-L1-L2-E7, TLR-8-HSP70-L1-L2-E7 and TLR-9-HSP70-L1-L2-E7 constructs was − 929.0, − 1064.0, − 974.3, − 1249.1, − 990.2, − 1034.0, respectively as shown in Fig. 12. The lowest energy levels determine the highest binding affinity between all of the predicted docked complexes.
In Vitro Experiments
Confirmation of the Plasmid DNA
At first, the pUC-57 vector harboring the HSP70-L1-L2-E7 fusion DNA construct (Fig. 13) was designed and synthesized. Then, the L1-L2-E7 and HSP70-L1-L2-E7 genes were successfully subcloned into pEGFP-N1 and pcDNA3.1 (-) eukaryotic vectors. After digestion, the L1-L2-E7 and HSP70-L1-L2-E7 genes with the clear bands of ~ 519 bp and ~ 753 bp were confirmed on agarose gel, respectively (Fig. 14).
Expression of the L1-L2-E7 and HSP70-L1-L2-E7 Genes In Vitro
The DNA constructs (pEGFP-L1-L2-E7 and pEGFP-HSP70-L1-L2-E7) were transfected into the eukaryotic cell line (HEK-293T) using TurboFect transfection reagent. The data showed that the cellular uptake of pEGFP-L1-L2-E7 and pEGFP-HSP70-L1-L2-E7 into the HEK-293T cells was ~ 56.16% ± 0.31 and ~ 80.45% ± 0.81, respectively. Moreover, the green cells were detected for the L1-L2-E7 and HSP70-L1-L2-E7 DNA delivery in HEK-293T cells using fluorescent microscopy (Fig. 15). The successful expression of the L1-L2-E7 and HSP70-L1-L2-E7 multiepitope peptides fused to GFP was confirmed by Western blotting, as well. Our results showed the clear bands of ~ 46.5, ~ 55.5 and ~ 27 kDa for the L1-L2-E7-GFP, HSP70-L1-L2-E7-GFP and GFP constructs, respectively (Fig. 16).
Evaluation of the GFP (B), L1-L2-E7-GFP (C) and HSP70-L1-L2-E7-GFP (D) DNA delivery into HEK-293T mammalian cells using TurboFect. Transfection efficiency was monitored by flow cytometry (right) and fluorescent microscopy (left) at 48 h post-transfection as compared to the untransfected cells as a negative control (A)
Identification of protein expression in HEK-293T cells using Western blotting: The clear bands were observed for GFP (lane 1, ~ 27 kDa), L1-L2-E7-GFP (lane 2, ∼ 46.5 kDa), and HSP70-L1-L2-E7-GFP (lane 3, ~ 55.5 kDa), respectively. No clear band was detected in untransfected cells as a negative control (lane 4). MW is molecular weight marker (prestained protein ladder, 10–180 kDa, Fermentas)
In Vivo Studies
Preventive Study
Antibody Assay
The levels of total IgG, IgG1 and IgG2a in mice immunized with the HSP70-L1-L2-E7 DNA construct (G2) was significantly higher than other groups against both L1-L2-E7 and HSP70-L1-L2-E7 multiepitope peptides as antigens (p < 0.05, Fig. 17). In addition, the levels of IgG2a in groups immunized with the HSP70-L1-L2-E7 (G2) and L1-L2-E7 (G1) DNA constructs were significantly higher than the levels of IgG1 especially against the HSP70-L1-L2-E7 multiepitope peptide as an antigen (p < 0.05). No significant antigen-specific antibody response was observed in the control sera.
Antibody responses against the L1-L2-E7 and HSP70-L1-L2-E7 peptides as an antigen in different groups: A total IgG, B IgG1, and C IgG2a; Mice sera were prepared from the whole blood samples of each group (n = 8) 3 weeks after the last immunization. All analyses were performed in duplicate for each sample shown as mean absorbance at 450 nm ± SD. ***p < 0.001; **p < 0.01; *p < 0.05; ns non-significant
Cytokine Detection
As detected, the levels of IFN-γ and IL-4 secretion in groups immunized with the L1-L2-E7 (G1) and HSP70-L1-L2-E7 (G2) DNA constructs were drastically higher than control groups (p < 0.05, Fig. 18). In addition, the injection of the HSP70-L1-L2-E7 (G2) DNA construct increased significantly IFN-gamma response compared to the injection of L1-L2-E7 (G1) DNA construct (p < 0.05). In contrast, there was no significant difference in IL-4 secretion between groups immunized with the HSP70-L1-L2-E7 and L1-L2-E7 DNA constructs (G1 & G2; p > 0.05). Furthermore, our results indicated that the ratios of IFN-γ/IL-4 were higher in test groups (G1 & G2) than control groups; therefore, they could induce Th1 immune response. On the other hand, no significant difference was observed in IL-10 secretion between different groups (test groups compared to control groups; p > 0.05, data not shown).
The levels of IFN-γ (A) and IL-4 (B) in vaccinated groups with different formulations: The pooled splenocytes were prepared from three mice in each group (n = 3) and re-stimulated with the L1-L2-E7 or HSP70-L1-L2-E7 peptides in vitro. The levels of cytokines were measured in the supernatant with ELISA as mean absorbance at 450 nm ± SD for each sample. All analyses were performed in duplicate for each sample. ***p < 0.001; **p < 0.01; *p < 0.05; ns non-significant
Granzyme B Secretion
The secretion of Granzyme B in groups immunized with the HSP70-L1-L2-E7 and L1-L2-E7 DNA constructs (G1 and G2) was considerably higher than the control groups (p < 0.001, Fig. 19). The group immunized with the HSP70-L1-L2-E7 DNA construct (G2) produced significantly higher levels of Granzyme B than other group (G1, p < 0.001) against both antigens.
Evaluation of Tumor Growth
To determine the preventive effects of the designed L1-L2-E7 and HSP70-L1-L2-E7 DNA constructs, tumor growth and survival rate were determined for 65 days after C3 challenge. The groups immunized with the L1-L2-E7 and HSP70-L1-L2-E7 DNA constructs (G1 & G2) indicated drastically lower tumor growth than that in control groups (G3 & G4, p < 0.01; Fig. 20A). Tumor growth was observed in control groups on approximately 7–14 days. The group vaccinated with the HSP70-L1-L2-E7 DNA construct (G2) reduced tumor growth more than the group immunized with the L1-L2-E7 DNA construct (G1) but it was not significant (p > 0.05; Fig. 20A). In addition, the group immunized with the HSP70-L1-L2-E7 DNA construct (G2) indicated a higher survival percentage (~ 80%) than the group immunized with the L1-L2-E7 DNA construct (G1; ~ 60%; Fig. 20B).
Preventive study A, B: Tumor growth curve and survival percentage in different groups: The mice were challenged with 1 × 105 C3 tumor cells 3 weeks after the last immunization. Tumor volumes were measured twice a week (A), and the percentage of tumor-free mice (or survival rate) was evaluated in different groups (B); Therapeutic study (C): Tumor growth curve; Tumor volumes were measured twice a week. ***p < 0.001; **p < 0.01; *p < 0.05; ns non-significant
Therapeutic Effects
Mice with the established tumors (~ 2–3 mm3) were treated by the L1-L2-E7 and HSP70-L1-L2-E7 DNA constructs. Among these groups, the group vaccinated by the HSP70-L1-L2-E7 DNA construct showed a higher survival rate (G2, ~ 80%) than the L1-L2-E7 DNA construct (G1, ~ 60%) and control groups (G3 & G4, 0%) 65 days following treatment. This result was similar to the preventive effects. Indeed, the tumor growth was stopped in 80% and 60% of the treated mice (G2 & G1, respectively). As shown in Fig. 20C, groups immunized with the DNA constructs (G1 & G2) indicated significantly lower tumor growth than that in control groups (G3 & G4, p < 0.01).
Discussion
The HPV L1/L2 capsid proteins and E7 oncoprotein are suitable targets for the prevention and treatment of cervical cancer, respectively. The design of DNA- or peptide-based vaccine constructs containing both T- and B-cell epitopes results in boosting the strength and durability of immune responses [11, 15, 65, 66]. HSPs have been utilized as a potent adjuvant in immunotherapy of cancers and infectious diseases. Moreover, constructing an antigen-HSP fusion provides a promising approach to enhance the potency of DNA-, protein- or peptide-based vaccines [29, 36, 67]. On the other hand, to overcome the low immunogenicity of the peptide-based vaccines, immunobioinformatics tools are beneficial in determining novel and potent antigenic epitopes [68, 69]. The immunogenicity of L1, L2 and E7 epitopes in peptide-based vaccines was evaluated in a wide-range of studies. Feltkamp et al. determined the HPV16 E7 49RAHYNIVTF57 sequence as an MHC-I binding epitope which triggers CTL-mediated responses [63, 70]. About 10 years later, Kawana et al. introduced HPV16 L2 peptide (108VEETSFIDAGAP120) as a highly immunogenic epitope [71]. Hitzeroth and Kwak showed that L2 DNA vaccination inhibited the growth of L2-expressing C3 tumor cells [72]. On the other hand, multivalent L1 DNA vaccines could induce strong cellular and humoral immune responses [73, 74]. Moreover, chimeric L1 vaccines harboring cross-neutralizing L2 peptides (e.g., 17QLYKTCKQAGTCPPDIIPKV36 epitope) were suggested as promising second-generation prophylactic HPV vaccine candidates [75]. In 2015, an immunogenic HPV16 L2 epitope (12RASATQLYKTCKQAGTCPPDIIPKVEGKTI41) was introduced into the adenovirus 5 (Ad5) hexon as a practical method of generating a protective HPV vaccine [76]. In 2018, two 9-mer epitopes of HPV58 E7 including QAQPATANY and SSDEDEIGL were found as the most potential B- and T-cell epitopes, respectively [77]. Tsang et al. proposed six immunogenic epitopes of HPV16 E6 and E7 proteins including 11KLPQLCTEL19, 72KISEYRHYC80, 90QQYNKPLCDL99 from E6 protein, and 11YMLDLQPET19, 7TLHEYMLDL15, 77RTLEDLLMGT86 from E7 protein [78].
Our group previously determined the immunogenic and conserved epitopes of L1 and L2 proteins among the high-risk HPV types with the population coverage rates of 95.55% and 96.33%, respectively in worldwide [20]. In addition, we determined the immunogenic epitopes of E5, E6 and E7 oncoproteins from HPV-16, -18, -31 and -45 using a two-stage immunoinformatics method [33, 34]. In this study, we used the same epitopes of L1, L2 and E7 obtained from our previous studies [20, 33, 34], and designed a L1-L2-E7 multiepitope peptide construct for further in silico studies (e.g., B-cell epitope prediction, physicochemical characteristics, secondary structure modeling, 3D structure prediction, 3D structure refinement and validation, and binding prediction to TLRs). On the other hand, different computational tools were utilized to design an additional novel multiepitope peptide construct based on the immunodominant T-cell epitopes of HSP70 linked to the L1-L2-E7 multiepitope peptide construct.
Some studies showed that the efficiency of DNA vaccines can be enhanced by the linkage of HPV16 E7 gene to HSP70 gene. For example, the fusion of Mycobacterium tuberculosis HSP70 (MtHSP70) to the modified HPV 16 E7 (mE7) gene stimulated a stronger E7-specific CD8+ T-cell response, and a more significant therapeutic effect against E7-expressing tumor cells than the E7 DNA construct in mice [79, 80]. Also, Jiang et al. generated a recombinant protein vaccine based on the fusion of HSP70 to a melanoma tumor antigen (Mage-a1). The Mage-a1-HSP70 fusion construct could significantly increase immune responses and antitumor effects against Mage-a1-expressing tumors as compared to the Mage-a1 protein, and the combination of Mage-a1 + HSP70 proteins [29]. In 2019, Matsui et al. identified twenty-nine HSP70-derived peptides (9-mers) bound to HLA-class I using peptide-binding experiments [81].
In our study, the prediction of MHC-I and MHC-II binding HSP70 epitopes was analyzed using IEDB, NetMHCpan 4.1, and NetMHCIIpan. Hence, T-cell epitopes with the highest binding affinity scores were selected. Generally, the MHC-II binding epitopes included HPV16 L1 (416DTYRYVQSQAITCQK430), HPV18 L1 (8DNTVYLPPPSVARVV22), HPV16 L2 (281 PDFLDIVALHRPALTSR297), HPV18 L2 (274SDFMDIIRLHRPALTSR290), HSP70 (168NVLRIINEPTAAAIA182), and HSP-70 (389QDLLLLDVAPLSLGL403). The MHC-I binding epitopes included HPV16 L1 (12YLPPVPVSKV21), HPV18 L1 (461DQYPLGRKFLV471), HPV16 L2 (11KRASATQLYK20), HPV18 L2 (11KRASVTDLYK20), HPV16 E7 (43GQAEPDRAHYNIVTF57), HPV18 E7 (78SSADDLRAFQQLFL91), HSP70 (113FYPEEISSMVLTKM126), and HSP70 (285SLFEGIDFYTSITR298).
The predicted HSP70 epitopes showed a high quality of proteasomal cleavage and Tap transport efficiency, as well. These epitopes were non-allergen using AlgPred server. In the next step, the population coverage rates for CTL and HTL epitopes were studied in sixteen-specified geographical regions for the predicted HSP70 epitopes. These data suggested a specific binding of the CTL and HTL epitopes to the prevalent HLA molecules in the targeted populations. For example, the highest world’s population coverage for two CTL epitopes of HSP70 was calculated about 88.39% and 81.19%, and for two HTL epitopes of HSP70 were determined about 40.07% and 21.86%. Furthermore, the MHC-I binding HSP70 epitopes (113–126 and 285–298 epitopes), and the MHC-II binding HSP70 epitopes (168–182 and 389–403 epitopes) showed the highest docking scores as compared to other HSP70 epitopes. In addition, all selected epitopes of L1, L2, E7 and HSP70 could induce IFN-gamma cytokine. As known, IFN-gamma has a major role in intracellular immunity against HPV infection [82]. Finally, the L1-L2-E7 and HSP70-L1-L2-E7 multiepitope peptide constructs were designed. In the next step, further immunoinformatics tools were applied to determine some physicochemical, structural and immunological properties of the designed constructs. The physicochemical properties of the L1-L2-E7 and HSP70-L1-L2-E7 constructs indicated that they were unstable and less soluble. In addition, the L1-L2-E7 construct with molecular weight 19.5 kDa and 173 amino acids had 20 positive charge residues (Arg + Lys) and 12 negative charge residues (Asp + Glu) and the HSP70-L1-L2-E7 construct with molecular weight 28.5 kDa and 255 amino acids consisted of 24 positive and 21 negative charge residues. The 3D modeling of the L1-L2-E7 and HSP70-L1-L2-E7 constructs using I-TASSER server showed that the C-scores of the best models for the L1-L2-E7 and HSP70-L1-L2-E7 constructs were − 3.66 and − 2.72, respectively. These data determined that the accuracy of the HSP70-L1-L2-E7 construct was higher than the L1-L2-E7 construct. The overall quality scores of the improved models for the L1-L2-E7 and HSP70-L1-L2-E7 constructs were 80.60, and 84.14, respectively after refinement. On the other hand, the molecular docking between the multiepitope peptide constructs and TLRs showed strong interactions between them using ClusPro server. In this line, several studies showed that simultaneous activation of multiple pathways of TLRs induced by vaccines led to stronger immunogenicity effects. Activation of TLRs signaling pathways could induce inflammatory reactions as well as promote DC maturation, HTL differentiation and production of an acquired immune response [83, 84]. The activation of TLRs by various ligands plays a major role in the development of cervical cancer. It was found that E7 oncoprotein can activate the PI3K/Akt/mTOR signaling pathway through TLRs in HPV-infected host epithelial cells. The expression levels of TLR2 and TLR4 were higher in cervical cancer cells than normal cells. In addition, TLR4, TLR5 and TLR9 were strongly related to HPV infection and cervical cancer. Thus, the TLR agonists were used for vaccine design in some studies [85,86,87].
Following the design of HSP70-L1-L2-E7 multiepitope peptide construct, it was reversely translated to the HSP70-L1-L2-E7 multiepitope DNA construct. After synthesis of the HSP70-L1-L2-E7 multiepitope gene in the cloning vector (pUC57), the L1-L2-E7 (∼ 519 bp) and HSP70-L1-L2-E7 (∼ 753 bp) genes were subcloned from pUC57-HSP70-L1-L2-E7 into pEGFP-N1 and pcDNA3.1 (-) eukaryotic expression vectors for in vitro and in vivo studies, respectively. For in vitro gene expression, the recombinant pEGFP-L1-L2-E7 and pEGFP-HSP70-L1-L2-E7 were transfected into HEK-293T cells using TurboFect reagent. The flow cytometry analysis showed that the L1-L2-E7 and HSP70-L1-L2-E7 genes were expressed about ~ 56.16% ± 0.31 and ~ 80.45% ± 0.81, respectively into the cells. Also, the expression of the L1-L2-E7 and HSP70-L1-L2-E7 multiepitope peptides fused to GFP was detected as the clear bands of ~ 46.5 and ~ 55.5 kDa, respectively by Western blotting.
The reports indicated that therapeutic HPV DNA vaccines possess effective antitumor activity. GX-188E is a HPV16/18 E6/E7 DNA therapeutic vaccine (Genexine, Inc.) achieved to phase II clinical trial. Moreover, VGX-3100 is a DNA vaccine containing two plasmids encoding the consensus HPV16/18 E6 and E7 genes achieved to phase III clinical trial [66, 88]. On the other hand, several strategies were used to increase the efficiency of DNA vaccines such as the use of various adjuvants (e.g., HSPs). Some studies focused on developing the effective DNA vaccines using different regions of HSPs (e.g., the N- or C-terminal fragments) in animal models [36]. Moreover, the multi-antigenic DNA constructs were used to boost immune responses, and generate both preventive and therapeutic effects. For example, a therapeutic HPV16 E7 DNA vaccine construct encoding the fusion sequence of the full-length bovin papillomavirus (BPV) L1 protein and a murine E7 antigenic epitope (aa 49–57) could induce effective E7-specific antitumor immune response in mice [89].
In our study, the endotoxin-free pcDNA-L1-L2-E7 and pcDNA-HSP70-L1-L2-E7 vectors were prepared and subcutaneously injected to C57BL/6 mice. Our results showed that the HSP70-L1-L2-E7 DNA construct generated significantly total IgG, IgG2a, IgG1, and IFN-gamma against both L1-L2-E7 and HSP70-L1-L2-E7 peptides as coated antigens compared to the L1-L2-E7 DNA construct. It is important that both DNA constructs elicited higher IgG2a and IFN-gamma levels than IgG1 and IL-4 levels indicating direction of responses toward Th1 response. In addition, the secretion of Granzyme B in the group vaccinated with pcDNA-HSP70-L1-L2-E7 was higher than the group vaccinated with pcDNA-L1-L2-E7 indicating an effective CTL activity in vitro. On the other hand, the percentage of tumor-free mice in the group vaccinated with pcDNA-HSP70-L1-L2-E7 was significantly more than the group vaccinated with pcDNA-L1-L2-E7. Moreover, the tumor growth in the group vaccinated with pcDNA-HSP70-L1-L2-E7 was less than the group vaccinated with pcDNA-L1-L2-E7, but this difference was not significant. Generally, the L1-L2-E7 DNA construct was effective for inducing immune responses and antitumor effects, but the linkage of HSP70 epitopes to the L1-L2-E7 DNA construct could significantly boost its potency.
Conclusion
In summary, two multiepitope peptide vaccine candidates were designed against HPV infection using in silico studies. After their reverse translation, four eukaryotic vectors expressing the multiepitope DNA (pEGFP-L1-L2-E7, pEGFP-HSP70-L1-L2-E7, pcDNA-L1-L2-E7 and pcDNA-HSP70-L1-L2-E7) were generated and used for in vitro and in vivo studies. The data showed that both multiepitope DNA constructs were successfully expressed in mammalian cell line. Moreover, in vivo studies showed that the linkage of HSP70 epitopes to the L1-L2-E7 DNA construct could significantly increase immune responses toward Th1 response and CTL activity and induce stronger antitumor effects. However, further studies are required to evaluate other strategies such as heterologous prime/boost regimen and/or the use of chemotherapeutic agents along with vaccination. Moreover, the stability of cytokine secretion will be assessed in splenocyte and plasma in future works.
References
Damania, B. (2016). A virological perspective on cancer. PLoS Pathogens, 12(2), e1005326.
Mesri, E. A., Feitelson, M. A., & Munger, K. (2014). Human viral oncogenesis: A cancer hallmarks analysis. Cell Host & Microbe, 15(3), 266–282.
Zapatka, M., Borozan, I., Brewer, D. S., Iskar, M., Grundhoff, A., Alawi, M., Desai, N., Sültmann, H., Moch, H., Cooper, C. S., Eils, R., Ferretti, V., & Lichter, P. (2020). The landscape of viral associations in human cancers. Nature Genetics, 52(3), 320–330.
Bravo, I. G., de Sanjosé, S., & Gottschling, M. (2010). The clinical importance of understanding the evolution of papillomaviruses. Trends in Microbiology, 18(10), 432–438.
McLaughlin-Drubin, M. E., & Munger, K. (2008). Viruses associated with human cancer. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease, 1782(3), 127–150.
Doorbar, J., Egawa, N., Griffin, H., Kranjec, C., & Murakami, I. (2015). Human papillomavirus molecular biology and disease association. Reviews in Medical Virology, 25, 2–23.
Doorbar, J., Quint, W., Banks, L., Bravo, I. G., Stoler, M., Broker, T. R., & Stanley, M. A. (2012). The biology and life-cycle of human papillomaviruses. Vaccine, 30, F55–F70.
De Villiers, E. M. (2013). Cross-roads in the classification of papillomaviruses. Virology, 445(1–2), 2–10.
Tsakogiannis, D., Gartzonika, C., Levidiotou-Stefanou, S., & Markoulatos, P. (2017). Molecular approaches for HPV genotyping and HPV-DNA physical status. Expert Reviews in Molecular Medicine, 19, e1.
Gupta, G., Glueck, R., & Patel, P. R. (2017). HPV vaccines: Global perspectives. Human Vaccines & Immunotherapeutics, 13(6), 1421–1424.
Wang, R., Pan, W., Jin, L., Huang, W., Li, Y., Wu, D., Gao, C., Ma, D., & Liao, S. (2020). Human papillomavirus vaccine against cervical cancer: Opportunity and challenge. Cancer Letters, 471, 88–102.
Ferlay, J., Colombet, M., Soerjomataram, I., Mathers, C., Parkin, D., Piñeros, M., Znaor, A., & Bray, F. (2019). Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. International Journal of Cancer, 144(8), 1941–1953.
Dadar, M., Chakraborty, S., Dhama, K., Prasad, M., Khandia, R., Hassan, S., Munjal, A., Tiwari, R., Karthik, K., Kumar, D., Iqbal, H. M. N., & Chaicumpa, W. (2018). Advances in designing and developing vaccines, drugs and therapeutic approaches to counter human papilloma virus. Frontiers in Immunology, 9, 2478.
Chabeda, A., Yanez, R. J., Lamprecht, R., Meyers, A. E., Rybicki, E. P., & Hitzeroth, I. I. (2018). Therapeutic vaccines for high-risk HPV-associated diseases. Papillomavirus Research, 5, 46–58.
Harper, D. M., & DeMars, L. R. (2017). HPV vaccines-a review of the first decade. Gynecologic Oncology, 146(1), 196–204.
Joura, E. A., Giuliano, A. R., Iversen, O. E., Bouchard, C., Mao, C., & Mehlsen, J. (2015). A 9-valent HPV vaccine against infection and intraepithelial neoplasia in women. New England Journal of Medicine, 372(8), 711–723.
Kim, H. J., & Kim, H. J. (2017). Current status and future prospects for human papillomavirus vaccines. Archives of Pharmacal Research, 40(9), 1050–1063.
Yang, A., Farmer, E., Wu, T. C., & Hung, C. F. (2016). Perspectives for therapeutic HPV vaccine development. Journal of Biomedical Science, 23(1), 75.
Yang, A., Jeang, J., Cheng, K., Cheng, T., Yang, B., Wu, T. C., & Hung, C. F. (2016). Current state in the development of candidate therapeutic HPV vaccines. Expert Review of Vaccines, 15(8), 989–1007.
Namvar, A., Bolhassani, A., Javadi, G., & Noormohammadi, Z. (2019). In silico/in vivo analysis of high-risk papillomavirus L1 and L2 conserved sequences for development of cross-subtype prophylactic vaccine. Scientific Reports, 9(1), 1–22.
Bahmani, B., Amini-bayat, Z., Ranjbar, M. M., Bakhtiari, N., & Zarnani, A. H. (2021). HPV16-E7 protein T cell epitope prediction and global therapeutic peptide vaccine design based on human leukocyte antigen frequency: An in silico study. International Journal of Peptide Research and Therapeutics, 27, 365–378.
Gomez-Gutierrez, J. G., Elpek, K. G., de Oca-Luna, R. M., Shirwan, H., Zhou, H. S., & McMasters, K. M. (2007). Vaccination with an adenoviral vector expressing calreticulin-human papillomavirus 16 E7 fusion protein eradicates E7 expressing established tumors in mice. Cancer Immunology, Immunotherapy, 56(7), 997–1007.
Reinis, M., Stepanek, I., Simova, J., Bieblova, J., Pribylova, H., & Indrova, M. (2010). Induction of protective immunity against MHC class I-deficient, HPV16-associated tumours with peptide and dendritic cell-based vaccines. International Journal of Oncology, 36(3), 545–551.
Milani, A., Basirnejad, M., & Bolhassani, A. (2019). Heat-shock proteins in diagnosis and treatment: An overview of different biochemical and immunological functions. Immunotherapy, 11(3), 215–239.
Bolhassani, A., & Rafati, S. (2008). Heat-shock proteins as powerful weapons in vaccine development. Expert Review of Vaccines, 7(8), 1185–1199.
Li, Y., Subjeck, J., Yang, G., Repasky, E., & Wang, X. Y. (2006). Generation of anti-tumor immunity using mammalian heat shock protein 70 DNA vaccines for cancer immunotherapy. Vaccine, 24(25), 5360–5370.
Udono, H., Ichiyanagi, T., Mizukami, S., & Imai, T. (2009). Heat shock proteins in antigen trafficking—Implications on antigen presentation to T cells. International Journal of Hyperthermia, 25(8), 617–625.
Binder, R. J., & Srivastava, P. K. (2005). Peptides chaperoned by heat-shock proteins are a necessary and sufficient source of antigen in the cross-priming of CD8+ T cells. Nature Immunology, 6(6), 593–599.
Jiang, J., Xie, D., Zhang, W., Xiao, G., & Wen, J. (2013). Fusion of Hsp70 to Mage-a1 enhances the potency of vaccine-specific immune responses. Journal of Translational Medicine, 11, 300.
Kaliamurthi, S., Selvaraj, G., Junaid, M., Khan, A., Gu, K., & Wei, D. Q. (2018). Cancer immunoinformatics: A promising era in the development of peptide vaccines for human papillomavirus-induced cervical cancer. Current Pharmaceutical Design, 24(32), 3791–3817.
Wei, D. Q., Selvaraj, G., & Kaushik, A. C. (2018). Computational perspective on the current state of the methods and new challenges in cancer drug discovery. Current Pharmaceutical Design, 24(32), 3725.
Kaliamurthi, S., Selvaraj, G., Kaushik, A. C., Gu, K. R., & Wei, D. Q. (2018). Designing of CD8+ and CD8+-overlapped CD4+ epitope vaccine by targeting late and early proteins of human papillomavirus. Biologics: Targets & Therapy, 12, 107.
Panahi, H. A., Bolhassani, A., Javadi, G., & Noormohammadi, Z. (2018). A comprehensive in silico analysis for identification of therapeutic epitopes in HPV16, 18, 31 and 45 oncoproteins. PLoS ONE, 13(10), e0205933.
Namvar, A., Panahi, H. A., Agi, E., & Bolhassani, A. (2020). Development of HPV 16, 18, 31, 45 E5 and E7 peptides-based vaccines predicted by immunoinformatics tools. Biotechnology Letters, 42(3), 403–418.
Negahdaripour, M., Nezafat, N., Heidari, R., Erfani, N., Hajighahramani, N., Ghoshoon, M. B., Shoolian, E., Rahbar, M. R., Najafipour, S., Dehshahri, A., Morowvat, M. H., & Ghasemi, Y. (2020). Production and preliminary in vivo evaluations of a novel in silico-designed L2-based potential HPV vaccine. Current Pharmaceutical Biotechnology, 21(4), 316–324.
Bolhassani, A., & Rafati, S. (2013). Mini-chaperones: Potential immuno-stimulators in vaccine design. Human Vaccines & Immunotherapeutics, 9(1), 153–161.
Kenter, G. G., Welters, M. J., Valentijn, A. R. P., Lowik, M. J., Berends-van der Meer, D. M., & Vloon, A. P. (2009). Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. New England Journal of Medicine, 361(19), 1838–18347.
Elhassan, R. M., Alsony, N. M., Othman, K. M., Izz-Aldin, D. T., Alhaj, T. A., Ali, A. A., Abashir, L. A., Ahmed, O. H., & Hassan, M. A. (2019). Computational vaccinology approach: Designing an efficient multi-epitope peptide vaccine against Cryptococcus neoformans var. grubii’s heat shock 70KDa protein. BioRxiv. https://doi.org/10.1101/534008
Jurtz, V., Paul, S., Andreatta, M., Marcatili, P., Peters, B., & Nielsen, M. (2017). NetMHCpan-4.0: Improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. The Journal of Immunology, 199(9), 3360–3368.
Jensen, K. K., Andreatta, M., Marcatili, P., Buus, S., Greenbaum, J. A., & Yan, Z. (2018). Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology, 154(3), 394–406.
Peters, B., Bulik, S., Tampe, R., Van Endert, P. M., & Holzhütter, H. G. (2003). Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. The Journal of Immunology, 171(4), 1741–1749.
Tenzer, S., Peters, B., Bulik, S., Schoor, O., Lemmel, C., Schatz, M., Kloetzel, P. M., Rammensee, H. G., Schild, H., & Holzhütter, H. G. (2005). Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cellular and Molecular Life Sciences, 62(9), 1025–1037.
Calis, J. J., Maybeno, M., Greenbaum, J. A., Weiskopf, D., De Silva, A. D., Sette, A., Kesmir, C., & Peters, B. (2013). Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Computational Biology, 9(10), e1003266.
Saha, S., & Raghava, G. (2006). AlgPred: Prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Research, 34(2), W202–W209.
Gupta, S., Kapoor, P., Chaudhary, K., Gautam, A., Kumar, R., & Raghava, G. P. (2013). In silico approach for predicting toxicity of peptides and proteins. PLoS ONE, 8(9), e73957.
Nagpal, G., Usmani, S. S., Dhanda, S. K., Kaur, H., Singh, S., Sharma, M., & Raghava, G. P. S. (2017). Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Scientific Reports, 7(1), 1–10.
Dhanda, S. K., Gupta, S., Vir, P., & Raghava, G. (2013). Prediction of IL-4 inducing peptides. Clinical and Developmental Immunology, 2013, 1–9.
Dhanda, S. K., Vir, P., & Raghava, G. P. (2013). Designing of interferon-γ inducing MHC class-II binders. Biology Direct, 8(1), 1–15.
Bui, H. H., Sidney, J., Dinh, K., Southwood, S., Newman, M. J., & Sette, A. (2006). Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics, 7(1), 1–5.
London, N., Raveh, B., Cohen, E., Fathi, G., & Schueler-Furman, O. (2011). Rosetta FlexPepDock web server-high resolution modeling of peptide-protein interactions. Nucleic Acids Research, 39(2), W249–W253.
Lee, H., Heo, L., Lee, M. S., & Seok, C. (2015). GalaxyPepDock: A protein–peptide docking tool based on interaction similarity and energy optimization. Nucleic Acids Research, 43(W1), W431–W435.
Jespersen, M. C., Peters, B., Nielsen, M., & Marcatili, P. (2017). BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Research, 45(W1), W24–W29.
Larsen, J. E. P., Lund, O., & Nielsen, M. (2006). Improved method for predicting linear B-cell epitopes. Immunome Research, 2(1), 1–7.
Gasteiger, E., Hoogland, C., Gattiker, A., Wilkins, M. R., Appel, R. D., & Bairoch, A. (2005). Protein identification and analysis tools on the ExPASy server (pp. 571–607). Springer.
Wang, S., Li, W., Liu, S., & Xu, J. (2016). RaptorX-Property: A web server for protein structure property prediction. Nucleic Acids Research, 44(W1), W430–W435.
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., & Zhang, Y. (2015). The I-TASSER Suite: Protein structure and function prediction. Nature Methods, 12(1), 7–8.
Lee, G. R., Heo, L., & Seok, C. (2016). Effective protein model structure refinement by loop modeling and overall relaxation. Proteins: Structure, Function, and Bioinformatics, 84, 293–301.
Lee, G. R., Heo, L., & Seok, C. (2018). Simultaneous refinement of inaccurate local regions and overall structure in the CASP12 protein model refinement experiment. Proteins: Structure, Function, and Bioinformatics, 86, 168–176.
Colovos, C., & Yeates, T. O. (1993). Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Science, 2(9), 1511–1519.
Vajda, S., Yueh, C., Beglov, D., Bohnuud, T., Mottarella, S. E., Xia, B., Hall, D. R., & Kozakov, D. (2017). New additions to the C lus P ro server motivated by CAPRI. Proteins: Structure, Function, and Bioinformatics, 85(3), 435–444.
Sabroe, I., Dower, S. K., & Whyte, M. K. (2005). The role of Toll-like receptors in the regulation of neutrophil migration, activation, and apoptosis. Clinical Infectious Diseases, 41(7), S421–S426.
Tartey, S., & Takeuchi, O. (2017). Pathogen recognition and Toll-like receptor targeted therapeutics in innate immune cells. International Reviews of Immunology, 36(2), 57–73.
Feltkamp, M. C., Smits, H. L., Vierboom, M. P., Minnaar, R. P., de Jongh, B. M., Drijfhout, J. W., Schegget, J. T., Melief, C. J., & Kast, W. M. (1993). Vaccination with cytotoxic T lymphocyte epitope-containing peptide protects against a tumor induced by human papillomavirus type 16-transformed cells. European Journal of Immunology, 23(9), 2242–2249.
Guruprasad, K., Reddy, B. B., & Pandit, M. W. (1990). Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Engineering, Design and Selection, 4(2), 155–161.
Melief, C. J., & Van Der Burg, S. H. (2008). Immunotherapy of established (pre) malignant disease by synthetic long peptide vaccines. Nature Reviews Cancer, 8(5), 351–360.
Rumfield, C. S., Roller, N., Pellom, S. T., Schlom, J., & Jochems, C. (2020). Therapeutic vaccines for HPV-associated malignancies. ImmunoTargets and Therapy, 9, 167–200.
Zong, J., Peng, Q., Wang, Q., Zhang, T., Fan, D., & Xu, X. (2009). Human HSP70 and modified HPV16 E7 fusion DNA vaccine induces enhanced specific CD8+ T cell responses and anti-tumor effects. Oncology Reports, 22(4), 953–961.
Oli, A. N., Obialor, W. O., Ifeanyichukwu, M. O., Odimegwu, D. C., Okoyeh, J. N., Emechebe, G. O., Adejumo, S. A., & Ibeanu, G. C. (2020). Immunoinformatics and vaccine development: An overview. ImmunoTargets and Therapy, 9, 13–30.
Soria-Guerra, R. E., Nieto-Gomez, R., Govea-Alonso, D. O., & Rosales-Mendoza, S. (2015). An overview of bioinformatics tools for epitope prediction: Implications on vaccine development. Journal of Biomedical Informatics, 53, 405–414.
Feltkamp, M. C., Vreugdenhil, G. R., Vierboom, M. P., Ras, E., van der Burg, S. H., Schegget, J. T., Melief, C. J. M., & Kast, W. M. (1995). Cytotoxic T lymphocytes raised against a subdominant epitope offered as a synthetic peptide eradicate human papillomavirus type 16-induced tumors. European Journal of Immunology, 25(9), 2638–2642.
Kawana, K., Yasugi, T., Kanda, T., Kino, N., Oda, K., & Okada, S. (2003). Safety and immunogenicity of a peptide containing the cross-neutralization epitope of HPV16 L2 administered nasally in healthy volunteers. Vaccine, 21(27–30), 4256–4260.
Hitzeroth, I. I., Passmore, J. A. S., Shephard, E., Stewart, D., Müller, M., Williamson, A. L., Rybicki, E. P., & Kast, W. M. (2009). Immunogenicity of an HPV-16 L2 DNA vaccine. Vaccine, 27(46), 6432–6434.
Kwak, K., Jiang, R., Jagu, S., Wang, J. W., Wang, C., Christensen, N. D., & Roden, R. B. S. (2013). Multivalent human papillomavirus l1 DNA vaccination utilizing electroporation. PLoS ONE, 8(3), e60507.
Lee, H. J., Yoon, J. K., Heo, Y., Cho, H., Cho, Y., Gwon, Y., Kim, K. C., Choi, J., Lee, J. S., Oh, Y. K., & Kim, Y. B. (2015). Therapeutic potential of an AcHERV-HPV L1 DNA vaccine. Journal of Microbiology, 53(6), 415–420.
McGrath, M., de Villiers, G. K., Shephard, E., Hitzeroth, I. I., & Rybicki, E. P. (2013). Development of human papillomavirus chimaeric L1/L2 candidate vaccines. Archives of Virology, 158(10), 2079–2088.
Wu, W. H., Alkutkar, T., Karanam, B., Roden, R. B., Ketner, G., & Ibeanu, O. A. (2015). Capsid display of a conserved human papillomavirus L2 peptide in the adenovirus 5 hexon protein: A candidate prophylactic hpv vaccine approach. Virology Journal, 12(1), 140.
Sabah, S. N., Gazi, M. A., Sthity, R. A., Husain, A. B., Quyyum, S. A., Rahman, M., & Islam, M. R. (2018). Designing of epitope-focused vaccine by targeting E6 and E7 conserved protein sequences: An immuno-informatics approach in human papillomavirus 58 isolates. Interdisciplinary Sciences: Computational Life Sciences, 10(2), 251–260.
Tsang, K. Y., Fantini, M., Fernando, R. I., Palena, C., David, J. M., Hodge, J. W., Gabitzsch, E. S., Jones, F. R., & Schlom, J. (2017). Identification and characterization of enhancer agonist human cytotoxic T-cell epitopes of the human papillomavirus type 16 (HPV16) E6/E7. Vaccine, 35(19), 2605–2611.
Chen, C. H., Wang, T. L., Hung, C. F., Yang, Y., Young, R. A., Pardoll, D. M., & Wu, T. C. (2000). Enhancement of DNA vaccine potency by linkage of antigen gene to an HSP70 gene. Cancer Research, 60(4), 1035–1042.
Zong, J., Wang, C., Wang, Q., Peng, Q., Xu, Y., Xie, X., & Xu, X. (2013). HSP70 and modified HPV 16 E7 fusion gene without the addition of a signal peptide gene sequence as a candidate therapeutic tumor vaccine. Oncology Reports, 30(6), 3020–3026.
Matsui, H., Hazama, S., Tamada, K., Udaka, K., Irie, A., Nishimura, Y., Miyakawa, T., Doi, S., Nakajima, M., Kanekiyo, S., Tokumitsu, Y., Shindo, Y., Tomochika, S., Yoshida, S., Iida, M., Suzuki, N., Takeda, S., Yamamoto, S., Yoshino, S., … Nagano, H. (2019). Identification of a promiscuous epitope peptide derived from HSP70. Journal of Immunotherapy, 42(7), 244.
Day, P. M., Thompson, C. D., Lowy, D. R., & Schiller, J. T. (2017). Interferon gamma prevents infectious entry of human papillomavirus 16 via an L2-dependent mechanism. Journal of Virology, 91(10), 1–10.
Theodoropoulos, G. E., Saridakis, V., Karantanos, T., Michalopoulos, N. V., Zagouri, F., Kontogianni, P., Lymperi, M., Gazouli, M., & Zografos, G. C. (2012). Toll-like receptors gene polymorphisms may confer increased susceptibility to breast cancer development. The Breast, 21(4), 534–538.
Dabbagh, K., & Lewis, D. B. (2003). Toll-like receptors and T-helper-1/T-helper-2 responses. Current Opinion in Infectious Diseases, 16(3), 199–204.
Yang, X., Cheng, Y., & Li, C. (2017). The role of TLRs in cervical cancer with HPV infection: A review. Signal Transduction and Targeted Therapy, 2(1), 1–10.
El-Omar, E., Ng, M., & Hold, G. (2008). Polymorphisms in toll-like receptor genes and risk of cancer. Oncogene, 27(2), 244–252.
de Matos, L. G., Cândido, E. B., Vidigal, P. V., Bordoni, P. H., Lamaita, R. M., Carneiro, M. M., & da Silva-Filho, A. L. (2017). Association between Toll-like receptor and tumor necrosis factor immunological pathways in uterine cervical neoplasms. Tumori Journal, 103(1), 81–86.
Choi, Y. J., Hur, S. Y., Kim, T. J., Hong, S. R., Lee, J. K., Cho, C. H., Park, K. S., Woo, J. W., Sung, Y. C., Suh, Y. S., & Park, J. S. (2020). A phase II, prospective, randomized, multicenter, open-label study of GX-188E, an HPV DNA vaccine, in patients with cervical intraepithelial neoplasia 3. Clinical Cancer Research, 26, 1616–1623.
Yang, A., Peng, S., Farmer, E., Zeng, Q., Cheng, M. A., Pang, X., Wu, T. C., & Hung, C. F. (2017). Enhancing antitumor immunogenicity of HPV16-E7 DNA vaccine by fusing DNA encoding E7-antigenic peptide to DNA encoding capsid protein L1 of Bovine papillomavirus. Cell & Bioscience, 7, 46.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest related to this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kayyal, M., Bolhassani, A., Noormohammadi, Z. et al. In Silico Design and Immunological Studies of Two Novel Multiepitope DNA-Based Vaccine Candidates Against High-Risk Human Papillomaviruses. Mol Biotechnol 63, 1192–1222 (2021). https://doi.org/10.1007/s12033-021-00374-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-021-00374-z