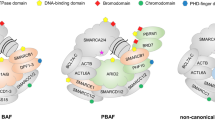
Abstract
Chromatin remodeling is a critical step in the DNA damage response, and the ATP-dependent chromatin remodelers are a group of epigenetic regulators that alter nucleosome assembly and regulate transcription factor accessibility to DNA, preventing genomic instability and tumorigenesis caused by DNA damage. The SWI/SNF chromatin remodeling complex is one of them, and mutations in the gene encoding the SWI/SNF subunit are frequently found in digestive tumors. We review the most recent literature on the role of SWI/SNF complexes in digestive tumorigenesis, with different SWI/SNF subunits playing different roles. They regulate the biological behavior of tumor cells, participate in multiple signaling pathways, interact with multiple genes, and have some correlation with the prognosis of patients. Their carcinogenic properties may help discover new therapeutic targets. Understanding the mutations and defects of SWI/SNF complexes, as well as the underlying functional mechanisms, may lead to new strategies for treating the digestive system by targeting relevant genes or modulating the tumor microenvironment.


Similar content being viewed by others
Data availability
The articles analyzed during the current study are available in the literature and listed in the references.
References
Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021. https://doi.org/10.3322/caac.21660.
Zheng RS, Zhang SW, Sun KX, et al. Cancer statistics in China, 2016. Zhonghua Zhong Liu Za Zhi. 2023. https://doi.org/10.3760/cma.j.cn112152-20220922-00647.
Wei W, Zeng H, Zheng R, et al. Cancer registration in China and its role in cancer prevention and control. Lancet Oncol. 2020. https://doi.org/10.1016/S1470-2045(20)30073-5.
Cao W, Chen H-D, Yu Y-W, et al. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl). 2021. https://doi.org/10.1097/CM9.0000000000001474.
Xia C, Dong X, Li H, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J. 2022. https://doi.org/10.1097/CM9.0000000000002108.
Nagtegaal ID, Odze RD, Klimstra D, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020. https://doi.org/10.1111/his.13975.
Jf F. WHO Classification of digestive tumors: the fourth edition. Ann Pathol. 2011. https://doi.org/10.1016/j.annpat.2011.08.001.
Kang MA, Lee J-S. A newly assigned role of CTCF in cellular response to broken DNAs. Biomolecules. 2021. https://doi.org/10.3390/biom11030363.
Ribeiro-Silva C, Vermeulen W, Lans H. SWI/SNF: Complex complexes in genome stability and cancer. DNA Repair. 2019. https://doi.org/10.1016/j.dnarep.2019.03.007.
Moison C, Chagraoui J, Caron M-C, et al. Zinc finger protein E4F1 cooperates with PARP-1 and BRG1 to promote DNA double-strand break repair. Proc Natl Acad Sci U S A. 2021. https://doi.org/10.1073/pnas.2019408118.
Vaicekauskaitė I, Sabaliauskaitė R, Lazutka JR, et al. The emerging role of chromatin remodeling complexes in ovarian cancer. IJMS. 2022. https://doi.org/10.3390/ijms232213670.
Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011. https://doi.org/10.1038/cr.2011.32.
Wang W, Côté J, Xue Y, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–82.
Pulice JL, Kadoch C. Composition and function of Mammalian SWI/SNF chromatin remodeling complexes in human disease. Cold Spring Harb Symp Quant Biol. 2016. https://doi.org/10.1101/sqb.2016.81.031021.
Mittal P, Roberts CWM. The SWI/SNF complex in cancer—biology, biomarkers and therapy. Nat Rev Clin Oncol. 2020. https://doi.org/10.1038/s41571-020-0357-3.
Romero OA, Sanchez-Cespedes M. The SWI/SNF genetic blockade: effects in cell differentiation, cancer and developmental diseases. Oncogene. 2014. https://doi.org/10.1038/onc.2013.227.
Mashtalir N, D’Avino AR, Michel BC, et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell. 2018. https://doi.org/10.1016/j.cell.2018.09.032.
Sima X, He J, Peng J, et al. The genetic alteration spectrum of the SWI/SNF complex: The oncogenic roles of BRD9 and ACTL6A. PLoS ONE. 2019. https://doi.org/10.1371/journal.pone.0222305.
Drage MG, Tippayawong M, Agoston AT, et al. Morphological features and prognostic significance of ARID1A-deficient esophageal adenocarcinomas. Arch Pathol Lab Med. 2017. https://doi.org/10.5858/arpa.2016-0318-OA.
Lowenthal BM, Nason KS, Pennathur A, et al. Loss of ARID1A expression is associated with DNA mismatch repair protein deficiency and favorable prognosis in advanced stage surgically resected esophageal adenocarcinoma. Hum Pathol. 2019. https://doi.org/10.1016/j.humpath.2019.09.004.
Schallenberg S, Bork J, Essakly A, et al. Loss of the SWI/SNF-ATPase subunit members SMARCF1 (ARID1A), SMARCA2 (BRM), SMARCA4 (BRG1) and SMARCB1 (INI1) in oesophageal adenocarcinoma. BMC Cancer. 2020. https://doi.org/10.1186/s12885-019-6425-3.
Zhou Z, Huang D, Yang S, et al. Clinicopathological significance, related molecular changes and tumor immune response analysis of the abnormal SWI/SNF complex subunit PBRM1 in gastric adenocarcinoma. Pathol Oncol Res. 2022. https://doi.org/10.3389/pore.2022.1610479.
Glückstein M-I, Dintner S, Arndt TT, et al. Comprehensive immunohistochemical study of the SWI/SNF complex expression status in gastric cancer reveals an adverse prognosis of SWI/SNF deficiency in genomically stable gastric carcinomas. Cancers (Basel). 2021. https://doi.org/10.3390/cancers13153894.
Zhu YP, Sheng LL, Wu J, et al. Loss of ARID1A expression is associated with poor prognosis in patients with gastric cancer. Hum Pathol. 2018. https://doi.org/10.1016/j.humpath.2018.04.003.
Shih-Chiang Huang, Chen K-H, Ng K-F, et al. Dedifferentiation-like tubular and solid carcinoma of the stomach shows phenotypic divergence and association with deficient SWI/SNF complex. Virchows Arch. 2022. https://doi.org/10.1007/s00428-022-03288-6
Tsuruta S, Kohashi K, Yamada Y, et al. Solid-type poorly differentiated adenocarcinoma of the stomach: Deficiency of mismatch repair and SWI/SNF complex. Cancer Sci. 2020. https://doi.org/10.1111/cas.14301.
Huang S, Ng K, Yeh T, et al. The clinicopathological and molecular analysis of gastric cancer with altered SMARCA4 expression. Histopathology. 2020. https://doi.org/10.1111/his.14117.
Huang S-C, Ng K-F, Chang IY-F, et al. The clinicopathological significance of SWI/SNF alterations in gastric cancer is associated with the molecular subtypes. PLoS One. 2021. https://doi.org/10.1371/journal.pone.0245356
Zhang Z, Li Q, Sun S, et al. Clinicopathological and prognostic significance of SWI/SNF complex subunits in undifferentiated gastric carcinoma. World J Surg Onc. 2022. https://doi.org/10.1186/s12957-022-02847-0.
Sasaki T, Kohashi K, Kawatoko S, et al. Tumor progression by epithelial-mesenchymal transition in ARID1A- and SMARCA4-aberrant solid-type poorly differentiated gastric adenocarcinoma. Virchows Arch. 2022. https://doi.org/10.1007/s00428-021-03261-9.
Mochizuki K, Kawai M, Odate T, et al. SMARCB1/INI1 is diagnostically useful in distinguishing α-fetoprotein-producing gastric carcinoma from hepatocellular carcinoma. Anticancer Res. 2018;38:6865.
Sen M, Wang X, Hamdan FH, et al. ARID1A facilitates KRAS signaling-regulated enhancer activity in an AP1-dependent manner in colorectal cancer cells. Clin Epigenetics. 2019. https://doi.org/10.1186/s13148-019-0690-5.
Yang Z, Huang D, Meng M, et al. BAF53A drives colorectal cancer development by regulating DUSP5-mediated ERK phosphorylation. Cell Death Dis. 2022. https://doi.org/10.1038/s41419-022-05499-w.
Ke S-B, Qiu H, Chen J-M, et al. MicroRNA-202-5p functions as a tumor suppressor in colorectal carcinoma by directly targeting SMARCC1. Gene. 2018. https://doi.org/10.1016/j.gene.2018.08.064.
Melloul S, Mosnier J-F, Masliah-Planchon J, et al. Loss of SMARCB1 expression in colon carcinoma. CBM. 2020. https://doi.org/10.3233/CBM-190287.
Wang J, Andrici J, Sioson L, et al. Loss of INI1 expression in colorectal carcinoma is associated with high tumor grade, poor survival, BRAFV600E mutation, and mismatch repair deficiency. Hum Pathol. 2016. https://doi.org/10.1016/j.humpath.2016.04.018.
Villatoro TM, Ma C, Pai RK. Switch/sucrose nonfermenting nucleosome complex–deficient colorectal carcinomas have distinct clinicopathologic features. Hum Pathol. 2020. https://doi.org/10.1016/j.humpath.2020.03.009.
Ahadi MS, Fuchs TL, Clarkson A, et al. SWI/SNF complex (SMARCA4, SMARCA2, INI1/SMARCB1) deficient colorectal carcinomas are strongly associated with microsatellite instability: An incidence study in 4508 colorectal carcinomas. Histopathology. 2022. https://doi.org/10.1111/his.14612.
Agaimy A, Daum O, Märkl B, et al. SWI/SNF Complex-deficient Undifferentiated/Rhabdoid carcinomas of the gastrointestinal tract: a series of 13 cases highlighting mutually exclusive loss of SMARCA4 and SMARCA2 and frequent Co-inactivation of SMARCB1 and SMARCA2. Am J Surg Pathol. 2016. https://doi.org/10.1097/PAS.0000000000000554.
Agaimy A, Daum O, Michal M, et al. Undifferentiated large cell/rhabdoid carcinoma presenting in the intestines of patients with concurrent or recent non-small cell lung cancer (NSCLC): clinicopathologic and molecular analysis of 14 cases indicates an unusual pattern of dedifferentiated metastases. Virchows Arch. 2021. https://doi.org/10.1007/s00428-021-03032-6.
Fang J-Z, Li C, Liu X-Y, et al. Hepatocyte-specific Arid1a deficiency initiates mouse steatohepatitis and hepatocellular carcinoma. PLoS ONE. 2015. https://doi.org/10.1371/journal.pone.0143042.
Midorikawa Y, Yamamoto S, Tatsuno K, et al. Accumulation of molecular aberrations distinctive to hepatocellular carcinoma progression. Can Res. 2020. https://doi.org/10.1158/0008-5472.CAN-20-0225.
Zhang F-K, Ni Q-Z, Wang K, et al. Targeting USP9X–AMPK axis in ARID1A-Deficient hepatocellular carcinoma. Cell Mol Gastroenterol Hepatol. 2022. https://doi.org/10.1016/j.jcmgh.2022.03.009.
Zhang L, Sun T, Wu X-Y, et al. Delineation of a SMARCA4-specific competing endogenous RNA network and its function in hepatocellular carcinoma. World J Clin Cases. 2022. https://doi.org/10.12998/wjcc.v10.i29.10501
Chen Z, Lu X, Jia D, et al. Hepatic SMARCA4 predicts HCC recurrence and promotes tumour cell proliferation by regulating SMAD6 expression. Cell Death Dis. 2018. https://doi.org/10.1038/s41419-017-0090-8.
Guerrero-Martínez JA, Reyes JC. High expression of SMARCA4 or SMARCA2 is frequently associated with an opposite prognosis in cancer. Sci Rep. 2018. https://doi.org/10.1038/s41598-018-20217-3.
Wang P, Song X, Cao D, et al. Oncogene-dependent function of BRG1 in hepatocarcinogenesis. Cell Death Dis. 2020. https://doi.org/10.1038/s41419-020-2289-3.
Hong SH, Son KH, Ha SY, et al. Nucleoporin 210 serves a key scaffold for SMARCB1 in liver cancer. Can Res. 2021. https://doi.org/10.1158/0008-5472.CAN-20-0568.
Cai X, Zhou J, Deng J, Chen Z. Prognostic biomarker SMARCC1 and its association with immune infiltrates in hepatocellular carcinoma. Cancer Cell Int. 2021. https://doi.org/10.1186/s12935-021-02413-w.
Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012. https://doi.org/10.1038/nature11547.
Okamura R, Kato S, Lee S, et al. ARID1A alterations function as a biomarker for longer progression-free survival after anti-PD-1/PD-L1 immunotherapy. J Immunother Cancer. 2020. https://doi.org/10.1136/jitc-2019-000438.
Botta GP, Kato S, Patel H, et al. SWI/SNF complex alterations as a biomarker of immunotherapy efficacy in pancreatic cancer. JCI Insight. 2021. https://doi.org/10.1172/jci.insight.150453.
Shain AH, Giacomini CP, Matsukuma K, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci U S A. 2012. https://doi.org/10.1073/pnas.1114817109.
Batcher E, Madaj P, Gianoukakis AG. Pancreatic neuroendocrine tumors. Endocr Res. 2011. https://doi.org/10.3109/07435800.2010.525085.
Han X, Chen W, Chen P, et al. Aberration of ARID1A is associated with the tumorigenesis and prognosis of sporadic nonfunctional pancreatic neuroendocrine tumors. Pancreas. 2020. https://doi.org/10.1097/MPA.0000000000001535.
Sotozono H, Kanki A, Yasokawa K, et al. Value of 3-T MR imaging in intraductal papillary mucinous neoplasm with a concomitant invasive carcinoma. Eur Radiol. 2022. https://doi.org/10.1007/s00330-022-08881-6.
Kimura Y, Fukuda A, Ogawa S, et al. ARID1A maintains differentiation of pancreatic ductal cells and inhibits development of pancreatic ductal adenocarcinoma in mice. Gastroenterology. 2018. https://doi.org/10.1053/j.gastro.2018.03.039.
Ferri-Borgogno S, Barui S, McGee AM, et al. Paradoxical role of AT-rich interactive domain 1A in restraining pancreatic carcinogenesis. Cancers (Basel). 2020. https://doi.org/10.3390/cancers12092695.
Tessier-Cloutier B, Schaeffer DF, Bacani J, et al. Loss of switch/sucrose non-fermenting complex protein expression in undifferentiated gastrointestinal and pancreatic carcinomas. Histopathology. 2020. https://doi.org/10.1111/his.14096.
Yamamoto T, Kohashi K, Yamada Y, et al. Relationship between cellular morphology and abnormality of SWI/SNF complex subunits in pancreatic undifferentiated carcinoma. J Cancer Res Clin Oncol. 2022. https://doi.org/10.1007/s00432-021-03860-8.
L L, Y L, Y G, et al. Potential roles of PBRM1 on immune infiltration in cholangiocarcinoma. Int J Clin Exp Pathol 2020;13(10):2661–2676.
Zheng S, Zhu Y, Zhao Z, et al. Liver fluke infection and cholangiocarcinoma: a review. Parasitol Res. 2017. https://doi.org/10.1007/s00436-016-5276-y.
Namjan A, Techasen A, Loilome W, et al. ARID1A alterations and their clinical significance in cholangiocarcinoma. PeerJ. 2020. https://doi.org/10.7717/peerj.10464.
Fukunaga Y, Fukuda A, Omatsu M, et al. Loss of Arid1a and Pten in pancreatic ductal cells induces Intraductal Tubulopapillary Neoplasm via the YAP/TAZ pathway. Gastroenterology. 2022. https://doi.org/10.1053/j.gastro.2022.04.020.
Wang W, Friedland SC, Guo B, et al. ARID1A, a SWI/SNF subunit, is critical to acinar cell homeostasis and regeneration and is a barrier to transformation and epithelial-mesenchymal transition in the pancreas. Gut. 2019. https://doi.org/10.1136/gutjnl-2017-315541.
Baldi S, Zhang Q, Zhang Z, et al. ARID1A downregulation promotes cell proliferation and migration of colon cancer via VIM activation and CDH1 suppression. J Cell Mol Med. 2022. https://doi.org/10.1111/jcmm.17590.
Niedermaier B, Sak A, Zernickel E, et al. Targeting ARID1A-mutant colorectal cancer: depletion of ARID1B increases radiosensitivity and modulates DNA damage response. Sci Rep. 2019. https://doi.org/10.1038/s41598-019-54757-z.
Dong X, Song S, Li Y, et al. Loss of ARID1A activates mTOR signaling and SOX9 in gastric adenocarcinoma-rationale for targeting ARID1A deficiency. Gut. 2022. https://doi.org/10.1136/gutjnl-2020-322660.
Zhu Y, Li K, Yan L, et al. miR-223-3p promotes cell proliferation and invasion by targeting Arid1a in gastric cancer. Acta Biochim Biophys Sin (Shanghai). 2020. https://doi.org/10.1093/abbs/gmz151.
Bala P, Singh AK, Kavadipula P, et al. Exome sequencing identifies ARID2 as a novel tumor suppressor in early-onset sporadic rectal cancer. Oncogene. 2021. https://doi.org/10.1038/s41388-020-01537-z.
Li R, Li Y, Qin H, et al. ACTL6A promotes the proliferation of esophageal squamous cell carcinoma cells and correlates with poor clinical outcomes. OTT Volume. 2021. https://doi.org/10.2147/OTT.S288807.
Wei Z, Xu J, Li W, et al. SMARCC1 enters the nucleus via KPNA2 and plays an oncogenic role in bladder cancer. Front Mol Biosci. 2022. https://doi.org/10.3389/fmolb.2022.902220.
Lüönd F, Sugiyama N, Bill R, et al. Distinct contributions of partial and full EMT to breast cancer malignancy. Dev Cell. 2021. https://doi.org/10.1016/j.devcel.2021.11.006.
Erfani M, Zamani M, Hosseini SY, et al. ARID1A regulates E-cadherin expression in colorectal cancer cells: a promising candidate therapeutic target. Mol Biol Rep. 2021. https://doi.org/10.1007/s11033-021-06671-9.
Tomihara H, Carbone F, Perelli L, et al. Loss of ARID1A promotes epithelial-mesenchymal transition and sensitizes pancreatic tumors to proteotoxic stress. Can Res. 2021. https://doi.org/10.1158/0008-5472.CAN-19-3922.
Huang L-Y, Zhao J, Chen H, et al. SCFFBW7-mediated degradation of Brg1 suppresses gastric cancer metastasis. Nat Commun. 2018. https://doi.org/10.1038/s41467-018-06038-y.
Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer. 2020. https://doi.org/10.1038/s41568-019-0216-7.
Sun D, Teng F, Xing P, et al. ARID1A serves as a receivable biomarker for the resistance to EGFR-TKIs in non-small cell lung cancer. Mol Med. 2021. https://doi.org/10.1186/s10020-021-00400-5.
Kim Y-B, Ahn JM, Bae WJ, et al. Functional loss of ARID1A is tightly associated with high PD-L1 expression in gastric cancer. Int J Cancer. 2019. https://doi.org/10.1002/ijc.32140.
Zhang S, Zhou Y-F, Cao J, et al. mTORC1 promotes ARID1A degradation and oncogenic chromatin remodeling in hepatocellular carcinoma. Can Res. 2021. https://doi.org/10.1158/0008-5472.CAN-21-0206.
Wang SC, Nassour I, Xiao S, et al. SWI/SNF component ARID1A restrains pancreatic neoplasia formation. Gut. 2019. https://doi.org/10.1136/gutjnl-2017-315490.
Cunningham R, Hansen CG. The Hippo pathway in cancer: YAP/TAZ and TEAD as therapeutic targets in cancer. Clin Sci (Lond). 2022. https://doi.org/10.1042/CS20201474.
Chang L, Azzolin L, Di Biagio D, et al. The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature. 2018. https://doi.org/10.1038/s41586-018-0658-1.
Hillmer RE, Link BA. The roles of hippo signaling transducers Yap and Taz in chromatin remodeling. Cells. 2019. https://doi.org/10.3390/cells8050502.
Araki O, Tsuda M, Omatsu M, et al. Brg1 controls stemness and metastasis of pancreatic cancer through regulating hypoxia pathway. Oncogene. 2023. https://doi.org/10.1038/s41388-023-02716-4.
Liu S, Cao W, Niu Y, et al. Single-PanIN-seq unveils that ARID1A deficiency promotes pancreatic tumorigenesis by attenuating KRAS-induced senescence. Elife. 2021. https://doi.org/10.7554/eLife.64204.
Guo B, Friedland SC, Alexander W, et al. Arid1a mutation suppresses TGF-β signaling and induces cholangiocarcinoma. Cell Rep. 2022. https://doi.org/10.1016/j.celrep.2022.111253.
Tokunaga R, Xiu J, Goldberg RM, et al. The impact of ARID1A mutation on molecular characteristics in colorectal cancer. Eur J Cancer. 2020. https://doi.org/10.1016/j.ejca.2020.09.006.
Kamori T, Oki E, Shimada Y, et al. The effects of ARID1A mutations on colorectal cancer and associations with PD-L1 expression by stromal cells. Cancer Reports. 2022. https://doi.org/10.1002/cnr2.1420.
Huang W, Li H, Shi X, et al. Characterization of genomic alterations in Chinese colorectal cancer patients. Jpn J Clin Oncol. 2021. https://doi.org/10.1093/jjco/hyaa182.
Kim SY, Shen Q, Son K, et al. SMARCA4 oncogenic potential via IRAK1 enhancer to activate Gankyrin and AKR1B10 in liver cancer. Oncogene. 2021. https://doi.org/10.1038/s41388-021-01875-6.
Yao B, Gui T, Zeng X, et al. PRMT1-mediated H4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling. Genome Med. 2021. https://doi.org/10.1186/s13073-021-00871-5.
He D-D, Shang X-Y, Wang N, et al. BRD4 inhibition induces synthetic lethality in ARID2-deficient hepatocellular carcinoma by increasing DNA damage. Oncogene. 2022. https://doi.org/10.1038/s41388-022-02176-2.
Wang R, Chen M, Ye X, et al. Role and potential clinical utility of ARID1A in gastrointestinal malignancy. Mutation Research/Reviews in Mutation Research. 2021. https://doi.org/10.1016/j.mrrev.2020.108360.
Hause RJ, Pritchard CC, Shendure J, et al. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016. https://doi.org/10.1038/nm.4191.
Li Y, Yang X, Zhu W, et al. SWI/SNF complex gene variations are associated with a higher tumor mutational burden and a better response to immune checkpoint inhibitor treatment: a pan-cancer analysis of next-generation sequencing data corresponding to 4591 cases. Cancer Cell Int. 2022. https://doi.org/10.1186/s12935-022-02757-x.
Peng L, Li J, Wu J, et al. A pan-cancer analysis of SMARCA4 alterations in human cancers. Front Immunol. 2021. https://doi.org/10.3389/fimmu.2021.762598.
Wei X-L, Wang D-S, Xi S-Y, et al. Clinicopathologic and prognostic relevance of ARID1A protein loss in colorectal cancer. World J Gastroenterol. 2014. https://doi.org/10.3748/wjg.v20.i48.18404.
Funding
This research was funded by The National Natural Science Foundation of China (81802474).
Author information
Authors and Affiliations
Contributions
Conceptualization, H.L., and J.Z.; Writing – Original Draft Preparation, H.L., X.Z.(Xin Zheng), and Y.Z.; Writing – Review & Editing, J.Z., H.L. and X.Z.(Xin Zheng),Y.Z. and X.Z.(Xiao Zhang). All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
We assure you this manuscript has not been published in part or whole or is under consideration for publication elsewhere in any language. All the authors have thoroughly studied the manuscript and approved its consent and submission to the "Medical Oncology" journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liang, H., Zheng, X., Zhang, X. et al. The role of SWI/SNF complexes in digestive system neoplasms. Med Oncol 41, 119 (2024). https://doi.org/10.1007/s12032-024-02343-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-024-02343-3