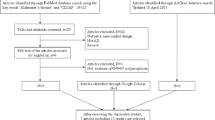
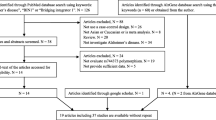
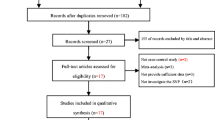
Abstract
Genome-wide association studies (GWAS) have associated several genetic variants with late-onset Alzheimer’s disease (LOAD), a neurodegenerative disease. Among those, rs3764650 ABCA7, rs6656401 CR1, and rs744373 BIN1 were associated as risk factors for LOAD, while rs11136000 CLU and rs610932 MS4A6A were protective. Recently, several case-control studies have investigated the association of these polymorphisms with AD. However, not all meta-analyses analyzed these variants across different ethnic groups. Therefore, we performed an updated meta-analysis of rs3764650 ABCA7, rs6656401 CR1, rs744373 BIN1, rs11136000 CLU, and rs610932 MS4A6A variants associated with LOAD, considering different ethnic populations. We utilized samples from 38 articles, comprising a total of 24,771 patients and 35,324 controls obtained through the PubMed database. Odds ratios (ORs) with 95% confidence intervals (CI) for polymorphisms were calculated by allelic comparison as an additive genetic model. We validated the risk for LOAD with BIN1 (rs744373), CR1 (rs6656401), and ABCA7 (rs376465), as well as the protective association for MS4A6A (rs610932) and CLU (rs11136000) variants.

Similar content being viewed by others
References
Bao J, Wang X, Mao Z (2016) Associations between genetic variants in 19p13 and 19q13 regions and susceptibility to Alzheimer disease: a meta-analysis. Med Sci Monit 22:234–243. https://doi.org/10.12659/MSM.895622
Chapuis J, Hansmannel F, Gistelinck M et al (2013) Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry 18:1225–1234. https://doi.org/10.1038/mp.2013.1
Chibnik LB, Shulman JM, Leurgans SE, Schneider JA, Wilson RS, Tran D, Aubin C, Buchman AS, Heward CB, Myers AJ, Hardy JA, Huentelman MJ, Corneveaux JJ, Reiman EM, Evans DA, Bennett DA, de Jager PL (2011) CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol 69:560–569. https://doi.org/10.1002/ana.22277
Condello C, Stohr J (2016) Aβ propagation and strains: implications for the phenotypic diversity in Alzheimer’s disease. Neurobiol Dis 109:191–200. https://doi.org/10.1016/j.nbd.2017.03.014
Deng YL, Liu LH, Wang Y, Tang HD, Ren RJ, Xu W, Ma JF, Wang LL, Zhuang JP, Wang G, Chen SD (2012) The prevalence of CD33 and MS4A6A variant in Chinese Han population with Alzheimer’s disease. Hum Genet 131:1245–1249. https://doi.org/10.1007/s00439-012-1154-6
Du W, Tan J, Xu W et al (2016) Association between clusterin gene polymorphism rs11136000 and late–onset alzheimer’s disease susceptibility: a review and meta–analysis of case–control studies. Exp Ther Med 12:2915–2927
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schürmann B, Heun R, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Hüll M, Rujescu D, Goate AM, Kauwe JSK, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, de Deyn PP, van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O'Donovan M, Owen MJ, Williams J (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 41:1088–1093. https://doi.org/10.1038/ng.440
Higgins JPT, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ Br Med J 327:557–560. https://doi.org/10.1136/bmj.327.7414.557
Hollingworth P, Harold D, Sims R et al (2011) Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43:429–435. https://doi.org/10.1038/ng.803
Ji W, Xu L, Zhou H, Wang S, Fang Y (2015) Meta-analysis of association between the genetic polymorphisms on chromosome 11q and Alzheimer’s disease susceptibility. Int J Clin Exp Med 8:18235–18244
Kadmiri N, Said N, Slassi I et al (2017) Biomarkers for Alzheimer disease: classical and novel candidates’ review. Neuroscience 370:181–190. https://doi.org/10.1016/j.neuroscience.2017.07.017
Kang S, Lee YH, Lee JE (2017) Metabolism-centric overview of the pathogenesis of Alzheimer’s disease. Yonsei Med J 58:479–488. https://doi.org/10.3349/ymj.2017.58.3.479
Karch CM, Goate AM (2015) Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry 77:43–51. https://doi.org/10.1016/j.biopsych.2014.05.006
Karch CM, Jeng AT, Nowotny P et al (2012) Expression of novel Alzheimer ’ s disease risk genes in control and Alzheimer ’ s disease brains. Plos One 7. https://doi.org/10.1371/journal.pone.0050976
Lambert JC, Amouyel P (2011) Genetics of Alzheimer’s disease: new evidences for an old hypothesis? Curr Opin Genet Dev 21:295–301. https://doi.org/10.1016/j.gde.2011.02.002
Lambert J-C, Heath S, Even G et al (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 41:1094–1099. https://doi.org/10.1038/ng.439
Lambert JC, Zelenika D, Hiltunen M, Chouraki V, Combarros O, Bullido MJ, Tognoni G, Fiévet N, Boland A, Arosio B, Coto E, Zompo MD, Mateo I, Frank-Garcia A, Helisalmi S, Porcellini E, Pilotto A, Forti P, Ferri R, Delepine M, Scarpini E, Siciliano G, Solfrizzi V, Sorbi S, Spalletta G, Ravaglia G, Valdivieso F, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossù P, Piccardi P, Annoni G, Seripa D, Galimberti D, Licastro F, Lathrop M, Soininen H, Amouyel P (2011) Evidence of the association of BIN1 and PICALM with the AD risk in contrasting European populations. Neurobiol Aging 32:756.e11–756.e15. https://doi.org/10.1016/j.neurobiolaging.2010.11.022
Liu G, Li F, Zhang S, Jiang Y, Ma G, Shang H, Liu J, Feng R, Zhang L, Liao M, Zhao B, Li K (2014a) Analyzing large-scale samples confirms the association between the ABCA7 rs3764650 polymorphism and Alzheimer’s disease susceptibility. Mol Neurobiol 50:757–764. https://doi.org/10.1007/s12035-014-8670-4
Liu G, Wang H, Liu J, Li J, Li H, Ma G, Jiang Y, Chen Z, Zhao B, Li K (2014b) The CLU gene rs11136000 variant is significantly associated with Alzheimer’s disease in Caucasian and Asian populations. NeuroMolecular Med 16:52–60. https://doi.org/10.1007/s12017-013-8250-1
Luo J, Li S, Qin X, Song L, Peng Q, Chen S, Xie Y, Xie L, Li T, He Y, Deng Y, Wang J, Zeng Z (2014) Meta-analysis of the association between CR1 polymorphisms and risk of late-onset Alzheimer’s disease. Neurosci Lett 578:165–170. https://doi.org/10.1016/j.neulet.2014.06.055
Ma X-Y, Yu J-T, Tan M-S et al (2013) Missense variants in CR1 are associated with increased risk of Alzheimer’ disease in Han Chinese. Neurobiol Aging 35:443.e17–443.e21. https://doi.org/10.1016/j.neurobiolaging.2013.08.009
Macaskill P, Walter SD, Irwig L (2001) A comparison of methods to detect publication bias in meta-analysis. Stat Med 20:641–654. https://doi.org/10.1002/sim.698
Mao YF, Guo ZY, Pu JL, Chen YX, Zhang BR (2015) Association of CD33 and MS4A cluster variants with Alzheimer’s disease in East Asian populations. Neurosci Lett 609:235–239. https://doi.org/10.1016/j.neulet.2015.10.007
Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, de Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JSK, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, George-Hyslop PS, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43:436–441. https://doi.org/10.1038/ng.801
Pant S, Sharma M, Patel K et al (2009) AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd in endocytic recycling. Cell 11:1399–1410. https://doi.org/10.1038/ncb1986.AMPH-1/Amphiphysin/Bin1
R Core Team (2011) R: A language and environment for statistical computing. R Fondation for Statistical Computing, Vienna. www.r-project.org
Ramirez LM, Goukasian N, Porat S, Hwang KS, Eastman JA, Hurtz S, Wang B, Vang N, Sears R, Klein E, Coppola G, Apostolova LG (2016) Common variants in ABCA7 and MS4A6A are associated with cortical and hippocampal atrophy. Neurobiol Aging 39:82–89. https://doi.org/10.1016/j.neurobiolaging.2015.10.037
Ren G, Vajjhala P, Lee JS, Winsor B, Munn AL (2006) The BAR domain proteins: molding membranes in fission, fusion, and phagy. Microbiol Mol Biol Rev 70:37–120. https://doi.org/10.1128/MMBR.70.1.37-120.2006
Rizzi F, Caccamo AE, Belloni L, Bettuzzi S (2009) Clusterin is a short half-life, poly-ubiquitinated protein, which controls the fate of prostate cancer cells. J Cell Physiol 219:314–323. https://doi.org/10.1002/jcp.21671
Rodrigues CL, Ziegelmann PK (2010) Metanálise: um guia prático. Rev HCPA 30:436–447
Rogers J, Li R, Mastroeni D, Grover A, Leonard B, Ahern G, Cao P, Kolody H, Vedders L, Kolb WP, Sabbagh M (2006) Peripheral clearance of amyloid beta peptide by complement C3-dependent adherence to erythrocytes. Neurobiol Aging 27:1733–1739. https://doi.org/10.1016/j.neurobiolaging.2005.09.043
Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Mizrachi I, Ostell J, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Yaschenko E, Ye J (2009) Database resources of the national center for biotechnology information. Nucleic Acids Res 37:D5–D15. https://doi.org/10.1093/nar/gkn741
Schmidt C, Wolff M, Von Ahsen N, Zerr I (2012) Alzheimer’s disease: genetic polymorphisms and rate of decline. Dement Geriatr Cogn Disord 33:84–89. https://doi.org/10.1159/000336790
Schwarzer G, Mair P, Hatzinger R (2007) Meta: an R package for meta-analysis. R News 7:40–45
Seshadri S (2010) Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303:1832–1840. https://doi.org/10.1001/jama.2010.574
Shen N, Chen B, Jiang Y, Feng R, Liao M, Zhang L, Li F, Ma G, Chen Z, Zhao B, Li K, Liu G (2014) An updated analysis with 85,939 samples confirms the association between CR1 rs6656401 polymorphism and Alzheimer’s disease. Mol Neurobiol 51:1017–1023. https://doi.org/10.1007/s12035-014-8761-2
Shulman JM, Chen K, Keenan BT, Chibnik LB, Fleisher A, Thiyyagura P, Roontiva A, McCabe C, Patsopoulos NA, Corneveaux JJ, Yu L, Huentelman MJ, Evans DA, Schneider JA, Reiman EM, de Jager PL, Bennett DA (2013) Genetic susceptibility for Alzheimer disease neuritic plaque pathology. JAMA Neurol 70:1150–1157. https://doi.org/10.1001/jamaneurol.2013.2815
Sterne JAC, Harbord RM (2004) Funnel plots in meta-analysis. Stata J 4:127–141
Sterne JAC, Sutton AJ, Ioannidis JPA et al (2011) Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ 342:d4002–d4002
Trikalinos TA, Salanti G, Khoury MJ, Ioannidis JPA (2006) Impact of violations and deviations in Hardy-Weinberg equilibrium on postulated gene-disease associations. Am J Epidemiol 163:300–309. https://doi.org/10.1093/aje/kwj046
Weis JH, Morton CC, Bruns GAP et al (1987) A complement receptor locus: genes encoding C3b/C4b receptor and C3d/Epstein-Barr virus receptor map to 1q32. J Immunol 138:312–315
Yu JT, Tan L, Hardy J (2014) Apolipoprotein E in Alzheimer’s disease: an update. Annu Rev Neurosci 37:79–100. https://doi.org/10.1146/annurev-neuro-071013-014300
Zhu R, Liu X, He Z (2016) The bridging integrator 1 gene polymorphism rs744373 and the risk of Alzheimer’s disease in Caucasian and Asian populations: an updated meta-analysis. Mol Neurobiol 54:1419–1428. https://doi.org/10.1007/s12035-016-9760-2
Acknowledgments
We value the assistance and technical support for research on the Núcleo de Genética Humana e Molecular – NGHM, Brazil.
Funding
This study was financially supported by Universidade Federal do Espírito Santo – UFES; Fundo de Amparo e Pesquisa do Espírito Santo – FAPES; Departamento de Ciência e Tecnologia do Ministério da Saúde – Decit; Secretaria de Ciência, Tecnologia e Insumos Estratégicos do Ministério da Saúde – SCTIE/MS; Fundo de Apoio à Ciência e Tecnologia do Município de Vitória – FACITEC; Ministério da Ciência, Tecnologia e Inovação – MCTI; Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPQ; Ministério da Educação – MEC; and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – CAPES.
Author information
Authors and Affiliations
Contributions
Jucimara Ferreira Figueiredo Almeida, Lígia Ramos dos Santos, Maira Trancozo, and Flavia de Paula wrote the manuscript and read and accepted the manuscript before submission. Jucimara Ferreira Figueiredo Almeida, Lígia Ramos dos Santos, and Maira Trancozo performed the meta-analysis, the literature search, and data extraction and evaluated the inclusion and exclusion criteria.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Fig. S1
(PPTX 437 kb)
Fig. S2
(PPTX 277 kb)
Fig. S3
(PPTX 138 kb)
Fig. S4
(PPTX 144 kb)
Fig. S5
(PPTX 73 kb)
Fig. S6
(PPTX 65 kb)
Fig. S7
(PPTX 57 kb)
Fig. S8
(PPTX 107 kb)
Fig. S9
(PPTX 96 kb)
Fig. S10
(PPTX 49 kb)
Fig. S11
(PPTX 96 kb)
Fig. S12
(PPTX 99 kb)
Fig. S13
(PPTX 325 kb)
Fig. S14
(PPTX 201 kb)
Fig. S15
(PPTX 82 kb)
Fig. S16
(PPTX 63 kb)
Fig. S17
(PPTX 60 kb)
Fig. S18
(PPTX 189 kb)
Fig. S19
(PPTX 199 kb)
Fig. S20
(PPTX 132 kb)
Fig. S21
(PPTX 179 kb)
Fig. S22
(PPTX 117 kb)
Fig. S23
(PPTX 131 kb)
Fig. S24
(PPTX 55 kb)
Fig. S25
(PPTX 70 kb)
Fig. S26
(PPTX 244 kb)
ESM 27
(DOCX 99 kb)
Rights and permissions
About this article
Cite this article
Almeida, J.F.F., dos Santos, L.R., Trancozo, M. et al. Updated Meta-Analysis of BIN1, CR1, MS4A6A, CLU, and ABCA7 Variants in Alzheimer’s Disease. J Mol Neurosci 64, 471–477 (2018). https://doi.org/10.1007/s12031-018-1045-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-018-1045-y