Abstract
Purpose
This was a review of circulating tumor DNA (ctDNA) in patients with peritoneal metastases from colorectal cancer.
Methods
We searched the PubMed database for studies reporting detection of ctDNA in patients with colorectal cancer (CRC) and with peritoneal metastases (PM) from colorectal cancer (CRPM). We extracted data on the population included, number of subjects, study design, type of ctDNA assay used and schedule, and the major findings from these publications.
Results
We identified 13 studies for review investigating ctDNA, using a variety of ctDNA assays, among 1787 patients with CRC without PM, as well as four eligible published and one unpublished (in press) studies, which included 255 patients with PM from any primary site and 61 patients with CRPM. Among the 13 studies investigating ctDNA among CRC without PM, posttreatment surveillance ctDNA was associated with recurrence and was generally more sensitive than imaging or tumor markers. Among the five studies including patients with PM, ctDNA was not universally able to detect the presence of PM, but when present, ctDNA predicted worse outcomes.
Conclusion
Circulating-tumor DNA is a potentially useful surveillance tool for patients with CRC. However, the sensitivity of ctDNA to detect CRPM is variable and warrants further inquiry.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is currently the third most common cancer in the USA with an annual incidence of approximately 150,000 [1]. It is the second most common cause of cancer-related deaths, contributing to approximately 52,000 annual deaths in the USA [1]. The peritoneum is a common site of metastasis, occurring in approximately 15% of patients with colorectal cancer [2]. The prognosis for patients with colorectal peritoneal metastases (CRPM) is poor, with a median survival of 12–16 months with systemic chemotherapy alone [3] and 42 months with surgical management in patients with isolated CRPM [4]. Visualization of CRPM is challenging with current imaging techniques, including computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET), which collectively have approximately an 85% sensitivity to detect peritoneal metastases (PM) [5]. As such, there remains significant treatment and diagnostic challenges with CRPM.
Cell-free DNA (cfDNA) is shed from both normal and diseased cells, most commonly those of hematopoietic origin. Furthermore, cfDNA is found in low concentrations within the bloodstream (1–10 ng/mL) and rapidly cleared by the liver (2.5-h half-life). DNA fragments released from tumor cells as circulating tumor DNA (ctDNA) are detectable in the bloodstream and comprise less than 0.1–10% of all circulating cfDNA [6]. Circulating tumor DNA is an emerging clinical tool for blood-based cancer screening, diagnosis, treatment guidance, and posttreatment surveillance [7].
Circulating tumor DNA assays vary by commercial availability, with each test having different gene numbers and types (somatic versus epigenetic) of alterations detected. A key difference is whether the analyzed genetic alterations are tumor tissue informed (defined from a patient’s tumor specimen) or tissue agnostic (extracted from a standardized alteration panel and gene methylation pattern) [7]. These variables considerably affect the intended use, sensitivity, and turnaround time of the ctDNA assay. Circulating tumor DNA has been investigated in multiple primary tumor types at multiple disease stages for the purpose of screening, detecting recurrence, surveying therapeutic response or resistance, and molecular profiling [8]. We sought to review the literature of ctDNA studies in CRC and in CRPM, specifically, as well as offer future directions for additional study.
Methods
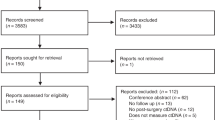
We performed a literature review by conducting a comprehensive search in PubMed/MEDLINE (through January 2023) using the following keywords: “circulating tumor DNA,” “ctDNA,” “colorectal cancer,” “peritoneal metastasis,” and “peritoneal carcinomatosis.” We included human studies published in English which analyzed ctDNA in CRC or peritoneal metastases (primarily CRPM) for surveillance or treatment guidance. Data extracted included the study population (stage of disease, proportion of patients with colorectal cancer in studies of peritoneal metastases), number of subjects, study design, type of ctDNA assay used and schedule, and the major findings from these publications.
Circulating Tumor DNA in Colorectal Cancer
The feasibility of ctDNA detection as a CRC tumor-burden assessment was first demonstrated by Diehl et al. in 2008 [9]. This study analyzed serial plasma samples for ctDNA in 18 subjects with stages II–IV colorectal cancer undergoing therapy and found detectable ctDNA was more highly associated with recurrence after surgical resection compared to blood carcinoembryonic antigen (CEA). A number of subsequent studies have further analyzed the use of ctDNA assays in colorectal cancer, primarily assessing for minimal/molecular residual disease (MRD) to determine the necessity of adjuvant therapy and predict risk of recurrence after definitive therapy.
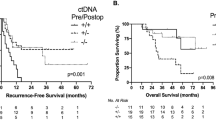
Several studies have been performed by Tie et al., using a ctDNA assay informed by the single tumor tissue alteration with the highest mean allele fraction (MAF, Safe-SeqS assay). This group initially demonstrated feasibility of ctDNA detection among 53 patients with metastatic CRC, and they found correlation of ctDNA levels with radiographic response and recurrence [10]. A subsequent study among 230 stage II colon cancer patients who underwent resection found 7.9% had detectable ctDNA postoperatively. Of those patients with positive ctDNA, 79% recurred versus only 9.8% of patients with negative ctDNA [11]. They also found an association between ctDNA detection and recurrence-free survival (RFS; HR 11, 95% CI, 1.8–68) among patients who received adjuvant chemotherapy. Another study among 159 patients with locally advanced rectal cancers found detection of ctDNA either post-chemoradiotherapy or post-resection was associated with worse RFS (HR 6.6, 95% CI, 2.6–17 and HR 13.0, 95% CI, 5.5–31, respectively) [12]. The 3-year RFS was 33% for patients with detectable postoperative ctDNA vs. 87% for postoperative ctDNA-negative patients. Furthermore, postoperative ctDNA status was an independent predictor of RFS on multivariate analysis. A stage III colon cancer study of 96 patients found an association between detectable postoperative ctDNA and post-adjuvant chemotherapy ctDNA and a shorter RFS (HR 3.8, 95% CI, 2.4–21.0 and HR 6.8, 95% CI 11.0–157.0, respectively). And again, postoperative ctDNA status was an independent predictor of RFS on multivariate analysis [13]. When evaluating the oligometastatic population, 54 patients undergoing resection for colorectal liver metastases found a ctDNA detection rate of 85% prior to surgery and 24% postoperatively [14]. Positive postoperative ctDNA was associated with shorter RFS (HR 6.3, 95% CI, 2.6–15.2) and OS (HR 4.2, 95% CI, 1.5–11.8) than those with negative postoperative ctDNA. End-of-treatment (postoperative and post-adjuvant therapy) ctDNA positivity had a 0% 5-year RFS versus 75.6% among those with negative end-of-treatment ctDNA (HR 14.9, 95% CI, 4.9–44.7).
Multiple additional single-arm studies have been performed investigating ctDNA for prognosis and surveillance in CRC. A Danish study of 130 patients with stages I-III CRC employing the tumor-informed ctDNA assay (Signatera) found (A) immediately postoperative, (B) post-adjuvant chemotherapy, and (C) surveillance detectable ctDNA was associated with higher rates of recurrence (HR 7.2, 95% CI, 2.7–19.0; HR 17.5, 95% CI, 5.4–56.5; and HR 43.5, 95% CI 9.8–193.5, respectively). Similar to previous studies, ctDNA remained independently associated with recurrence on multivariate analysis [15]. For those with radiographic recurrence, ctDNA detection preceded imaging findings of recurrence by a median of 8.7 months. A different study utilizing a tumor-informed orthogonal digital droplet PCR (ddPCR) ctDNA assay also found postoperative and surveillance ctDNA positivity were associated with disease-free survival (DFS; HR 6.96, 95% CI 2.57–18.91 and HR 8.03, 95% CI 1.79–35.98, respectively), and postoperative and serial ctDNA positivity remained a significant risk factor for DFS on multivariate analysis [16]. In the ddPCR ctDNA analysis, ctDNA detection preceded radiologic relapse by a median of 11.5 months. A second study using the ddPCR ctDNA assay among 29 patients with stages II–III rectal cancer found postoperative ctDNA positivity was also associated with PFS (HR 11.56, p = 0.007) [17].
In a study of 103 patients with stages I–IV CRC, a plasma-only (tumor tissue-agnostic) ctDNA assay incorporating genomic and epigenomic alterations (Guardant Reveal) was investigated [18]. Among patients with at least 1 year follow-up and with ctDNA-positive post-definitive treatment (postoperative and post-adjuvant therapy, if given), 100% recurred versus 24% of those with negative post-definitive treatment ctDNA (HR 11.2, p < 0.0001). The sensitivity of ctDNA to detect recurrence increased when longitudinal surveillance specimens were included (55.6 to 69.0%). A more recent study among 112 patients with metastatic, KRAS-mutant CRC compared two ctDNA assays: the Signatera tumor-informed assay and a KRAS alteration ctDNA panel [19]. Positive detection of postoperative ctDNA by the tumor-informed assay was associated with worse DFS (HR 5.8, 95% CI 3.5–9.7) and OS (HR 16.0, 95% CI 3.9–68) than those with undetectable postoperative ctDNA. Circulating tumor DNA remained a significant risk factor for worse DFS on multivariate analysis (HR 5.78, 95% CI 3.34–10.0). No stratification by metastatic site was performed. When comparing the two ctDNA assays, investigators found 44.5% discordance, with all discordant cases having undetectable ctDNA by KRAS panel but positive by the 16-alteration tumor-informed panel. Furthermore, 91.6% of the discordant cases recurred, suggesting higher sensitivity of the 16 gene tumor-informed ctDNA assay than the KRAS alteration assay. Another multicenter study using the 16 gene tumor-informed ctDNA assay in 168 patients with stage III CRC found postoperative (HR 7.0, 95% CI 3.7–13.5) and post-adjuvant therapy surveillance (HR 50.76, 95% CI 15.4–167) ctDNA detection predicted RFS [20]. The only patients with detectable postoperative ctDNA who did not recur were those who cleared ctDNA permanently after adjuvant therapy. The lead time for ctDNA versus radiologic detection of recurrence was a median of 9.8 months.
A recent retrospective study among 48 patients with stages II–IV CRC compared the surveillance sensitivity of the 16 gene tumor-informed ctDNA assay to imaging plus traditional carcinoembryonic antigen (CEA) for recurrence and found that ctDNA did not improve the sensitivity compared to imaging plus CEA (53.3% vs. 60.0% sensitivity, p > 0.99). Furthermore, ctDNA did not detect recurrence earlier than imaging plus CEA (14.3 months vs. 15.0 months) [21]. These findings, from a relatively small retrospective and single-center study, are in contrast to the studies above but highlight the notion that surveillance ctDNA in CRC may not be an actionable finding beyond imaging and standard of care blood biomarkers alone [22].
There has been one published prospective randomized controlled trial investigating ctDNA-guided adjuvant therapy in CRC [23]. This study, performed by Tie et al., randomized 455 patients with stage 2 CRC to ctDNA-guided adjuvant therapy (using the Safe-SeqS tumor-informed alteration assay) versus standard of care (clinicopathologic)-guided adjuvant therapy. They found 15% of patients in the ctDNA arm had detectable ctDNA and received adjuvant chemotherapy versus 28% receiving adjuvant therapy in the standard-management group. The ctDNA-guided group had a noninferior 2-year RFS compared to the standard-management group (93.5% v. 92.4%, absolute difference 1.1%, 95% CI − 4.1–6.2). The 3-year RFS was 86.4% among ctDNA-positive patients who received chemotherapy and 92.5% among those without detectable ctDNA who did not receive adjuvant chemotherapy Table 1 .
Circulating Tumor DNA from Peritoneal Metastases
The detectability of ctDNA from primary and metastatic peritoneal sites remains challenging. One challenge is that ctDNA from peritoneal disease may not enter the vasculature, limiting blood-based gene assays. The second issue is that peritoneal metastases may generate lower levels of ctDNA than other metastatic sites [6]. The rationale for lower detection levels of ctDNA from peritoneal metastases are uncertain but may include lower and more disorganized vascular density, inhibition by the plasma-peritoneal barrier, and mucin production disrupting ctDNA vascular entry [24,25,26].
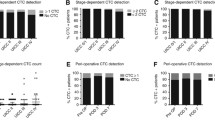
We have previously investigated perioperative ctDNA among patients undergoing surgery for colorectal peritoneal metastases (CRPM) and found approximately 63% had detectable preoperative and postoperative ctDNA, using a 73-gene tissue-agnostic panel (Guardant360). Importantly, those with high levels of preoperative or postoperative detectable ctDNA had worse PFS [27, 28]. There was high concordance between preoperative ctDNA and tissue DNA genomic alterations [29].
Another study investigated ctDNA analysis among 30 patients undergoing curative-intent cytoreductive surgery and hyperthermic intraperitoneal chemotherapy (CRS-HIPEC) for CRPM [30]. They used a 3-gene alteration tumor-informed ctDNA assay and found that 33% of patients had detectable ctDNA preoperatively which was associated with reduced DFS. In the post-CRS-HIPEC cases, only 4% (one patient) had detectable ctDNA, and this patient had radiographic distant recurrence 7 months later.
A larger retrospective study of 279 patients with stages II–IV gastrointestinal cancers investigated pretreatment ctDNA using a tumor-agnostic panel (Gaurdant360) and found that higher rates of detectable ctDNA correlated with higher stages of disease. There was a lower maximum variant allele frequency (mVAF) in those with peritoneal metastases than in those with non-peritoneal metastases (although the peritoneal metastasis patients had 72.1% ctDNA detection rate and the results were not stratified by primary tumor site) [31]. The authors concluded that “caution is warranted when interpreting ctDNA results” from patients with peritoneal metastases.
Recently, Singh et al. retrospectively evaluated the 16 gene tumor-informed assay in the clinical management of 13 patients with radiographically occult but pathologically confirmed peritoneal metastases (eight appendiceal and five gastric cancers) [32]. Detectable tumor-informed ctDNA was identified in 62% (8/13) of patients: three appendiceal (50% of cases) and five gastric (100% of cases) cancers. Three appendiceal patients had negative ctDNA, but detectable occult disease on laparoscopic analysis and two appendiceal cancer patients were unable to create a baseline test due to insufficient tissue. Even with low detectable ctDNA within the blood, disease burden correlated with longitudinal ctDNA analysis. Furthermore, nine of 13 patients had no other blood biomarker to follow their disease, whereas ctDNA was informative.
Future Directions of ctDNA in CRPM
Circulating tumor DNA has potential for risk stratification, surveillance for recurrence, and identification of actionable genetic alterations in patients with cancer. Patients with CRC have shown a significant association with ctDNA and RFS across multiple studies. It typically precedes imaging findings of recurrence by 8–12 months. A major unanswered question, however, is whether earlier detection of recurrence by ctDNA improves outcomes and if interventions should occur at ctDNA detection. There are cases of patients with increasing ctDNA without radiographic disease of progression that later have a reduction of ctDNA without intervention. It is unknown if a flood of cfDNA from another source mimics the ctDNA of the tumor, if a tumor cell indeed developed but immune surveillance responded appropriately, or if the assay resulted a true–false positive. Although not proven in solid tumor clinical trials, there is a rationale for focusing treatment efforts on minimal residual disease to improve oncologic outcomes [33]. Radiologic occult disease is of highest concern, especially in terms of peritoneal dissemination without definable or measurable masses. The disease burden within the abdomen can cause obstruction complications without early intervention. Furthermore, it is theorized that earlier disease stages may have more intact immune surveillance and potential responses versus those with later stage disease [34, 35]. As such, with less toxic and more effective treatments in low-burden disease states (i.e., immunotherapy), attempting treatment at the earliest sign of progression with ctDNA may provide improved quality of life and progression-free survival above lead time bias. Already, ctDNA has been shown to be predictive of immunotherapy responses in patients with solid tumors before radiologic response [36]. Withholding adjuvant therapy in patients who are ctDNA-negative post-resection in non-metastatic, low-risk CRC does not appear to cause harm [23], and additional studies are underway to determine how ctDNA might alter adjuvant treatment decisions in CRC [37].
Use of ctDNA in patients with CRPM for risk stratification has not been well-studied. Questions remain regarding the sensitivity of ctDNA assays in these patients. While some assays allow for identification of actionable genetic alterations to guide systemic therapy, this may be better determined through tumor tissue analysis than blood-based methods. Considering its paucity in the bloodstream, tumor-informed, multiple gene ctDNA assays provide higher sensitivity for peritoneal disease versus single, fixed, or droplet PCR ctDNA assays. Utilization of ctDNA for surveillance in patients with CRPM remains uncertain, and further research is needed to compare sensitivity of various ctDNA assays, as well as to determine the optimal timing and frequency of ctDNA analysis in this population. Furthermore, if blood is not an adequate milieu to evaluate ctDNA, peritoneal washings or ascites may be a more accurate source of ctDNA in CRPM patients, although this is not always available. Similar to nonmetastatic CRC, it also is critical to determine whether earlier detection of recurrence or progression by ctDNA and subsequent interventions in CRPM patients improves oncologic outcomes and quality of life.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33.
Data from: Surveillance, Epidemiology, and End Results (SEER) program. 2021;SEER*Stat Database: Incidence - SEER Research Data, Nov 2021 Sub (1975–2019). National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch. Deposited April 2022.
Franko J, Shi Q, Meyers JP, et al. Prognosis of patients with peritoneal metastatic colorectal cancer given systemic therapy: an analysis of individual patient data from prospective randomised trials from the Analysis and Research in Cancers of the Digestive System (ARCAD) database. Lancet Oncol. 2016;17(12):1709–19.
Quénet F, Elias D, Roca L, et al. Cytoreductive surgery plus hyperthermic intraperitoneal chemotherapy versus cytoreductive surgery alone for colorectal peritoneal metastases (PRODIGE 7): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2021;22(2):256–66.
Laghi A, Bellini D, Rengo M, et al. Diagnostic performance of computed tomography and magnetic resonance imaging for detecting peritoneal metastases: systematic review and meta-analysis. Radiol Med. 2017;122(1):1–15.
Bando H, Nakamura Y, Taniguchi H, et al. Effects of metastatic sites on circulating tumor DNA in patients with metastatic colorectal cancer. JCO Precis Oncol. 2022;6: e2100535.
Alese OB, Cook N, Ortega-Franco A, Ulanja MB, Tan L, Tie J. Circulating tumor DNA: an emerging tool in gastrointestinal cancers. Am Soc Clin Oncol Educ Book. 2022;42:1–20.
Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med. 2018;379(18):1754–65.
Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–90.
Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015;26(8):1715–22.
Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346ra92.
Tie J, Cohen JD, Wang Y, et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut. 2019;68(4):663–71.
Tie J, Cohen JD, Wang Y, et al. Circulating Tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 2019;5(12):1710–7.
Tie J, Wang Y, Cohen J, et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: a prospective cohort study. PLoS Med. 2021;18(5): e1003620.
Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5(8):1124–31.
Tarazona N, Gimeno-Valiente F, Gambardella V, et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann Oncol. 2019;30(11):1804–12.
McDuff SGR, Hardiman KM, Ulintz PJ, et al. Circulating tumor DNA predicts pathologic and clinical outcomes following neoadjuvant chemoradiation and surgery for patients with locally advanced rectal cancer. JCO Precis Oncol.
Parikh AR, Van Seventer EE, Siravegna G, et al. Minimal residual disease detection using a plasma-only circulating tumor DNA assay in patients with colorectal cancer. Clin Cancer Res. 2021;27(20):5586–94.
Loupakis F, Sharma S, Derouazi M, et al. Detection of molecular residual disease using personalized circulating tumor DNA assay in patients with colorectal cancer undergoing resection of metastases. JCO Precis Oncol. 2021;5:PO.21.00101.
Henriksen TV, Tarazona N, Frydendahl A, et al. Circulating tumor DNA in stage III colorectal cancer, beyond minimal residual disease detection, toward assessment of adjuvant therapy efficacy and clinical behavior of recurrences. Clin Cancer Res. 2022;28(3):507–17.
Fakih M, Sandhu J, Wang C, et al. Evaluation of comparative surveillance strategies of circulating tumor DNA, imaging, and carcinoembryonic antigen levels in patients with resected colorectal cancer. JAMA Netw Open. 2022;5(3):e221093–e221093.
Venook AP. Colorectal cancer surveillance with circulating tumor DNA assay. JAMA Netw Open. 2022;5(3):e221100–e221100.
Tie J, Cohen JD, Lahouel K, et al. Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer. N Engl J Med. 2022;386(24):2261–72.
Jacquet P, Sugarbaker PH. Peritoneal-plasma barrier. Cancer Treat Res. 1996;82:53–63.
Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24.
Kastelein AW, Vos LMC, van Baal J, et al. Poor perfusion of the microvasculature in peritoneal metastases of ovarian cancer. Clin Exp Metastasis. 2020;37(2):293–304.
Baumgartner JM, Riviere P, Lanman RB, et al. Prognostic utility of pre- and postoperative circulating tumor DNA liquid biopsies in patients with peritoneal metastases. Ann Surg Oncol. 2020;27(9):3259–67.
Baumgartner JM, Tobin L, Heavey SF, Kelly KJ, Roeland EJ, Lowy AM. Predictors of progression in high-grade appendiceal or colorectal peritoneal carcinomatosis after cytoreductive surgery and hyperthermic intraperitoneal chemotherapy. Ann Surg Oncol. 2015;22(5):1716–21.
Baumgartner JM, Raymond VM, Lanman RB, et al. Preoperative circulating tumor DNA in patients with peritoneal carcinomatosis is an independent predictor of progression-free survival. Ann Surg Oncol. 2018;25(8):2400–8.
Beagan JJ, Sluiter NR, Bach S, et al. Circulating tumor DNA as a preoperative marker of recurrence in patients with peritoneal metastases of colorectal cancer: a clinical feasibility study. J Clin Med. 2020;9(6):1738.
Sullivan BG, Lo A, Yu J, et al. Circulating tumor DNA is unreliable to detect somatic gene alterations in gastrointestinal peritoneal carcinomatosis. Ann Surg Oncol. 2023;30(1):278–84.
Singh H, Klempner SJ, Melnitchouk N, et al. Highly sensitive ctDNA assay aids clinical management of radiographically occultisolated peritoneal metastases in patients with gastrointestinal cancer. JCO Precis Oncol. In press.
Luskin MR, Murakami MA, Manalis SR, Weinstock DM. Targeting minimal residual disease: a path to cure? Nat Rev Cancer. 2018;18(4):255–63.
Cercek A, Lumish M, Sinopoli J, et al. PD-1 blockade in mismatch repair-deficient, locally advanced rectal cancer. N Engl J Med. 2022;386(25):2363–76.
Chalabi M. Defying all odds in MMR-deficient rectal cancers. Cancer Cell. 2022;40(9):914–6.
Bratman SV, Yang SYC, Iafolla MAJ, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer. 2020;1(9):873–81.
Taniguchi H, Nakamura Y, Kotani D, et al. CIRCULATE-Japan: circulating tumor DNA-guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Sci. 2021;112(7):2915–20.
Author information
Authors and Affiliations
Contributions
JB performed the initial literature review, wrote the manuscript text, prepared the table, and approved the final manuscript. GB performed additional literature review, provided critical revisions, and reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
Author G. B. is a consultant and advisory board member for Natera and TumorGen. The other author declares no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Baumgartner, J.M., Botta, G.P. Role of Circulating Tumor DNA Among Patients with Colorectal Peritoneal Metastases. J Gastrointest Canc 55, 41–46 (2024). https://doi.org/10.1007/s12029-023-00959-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-023-00959-8