Abstract
Purpose
Colorectal cancer (CRC) is a main cause of morbidity and mortality in the world. Chemoradioresistance is a major problem in CRC treatment. Identification of novel therapeutic targets in order to overcome treatment resistance in CRC is necessary.
Methods
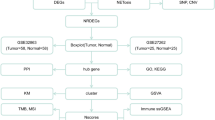
In this study, gene expression omnibus (GEO) database was searched to find microarray datasets. Data normalization/analyzing was performed using ExAtlas. The gene ontology (GO) and pathway enrichment analysis was performed using g:Profiler. Protein–protein interaction network (PPIN) was constructed by Search Tool for the Retrieval of Interacting Genes (STRING) and analyzed using Cytoscape. Survival analysis was done using Kaplan–Meier curve method.
Results
Forty-one eligible datasets were included in study. A total of 12,244 differentially expressed genes (DEGs) and 7337 unique DEGs were identified. Among them, 1187 DEGs were overlapped in ≥ 3 datasets. Fifty-five overlapped genes were considered as hub genes. Common hub genes in chemo/radiation/chemoradiation datasets were chosen as the essential candidate genes (n = 13). Forty-one hub gene and 7 essential candidate genes were contributed in the significant modules. The modules were mainly enriched in the signaling pathways of senescence, autophagy, NF-κB, HIF-1, stem cell pluripotency, notch, neovascularization, cell cycle, p53, chemokine, and PI3K-Akt. NGFR, FGF2, and PROM1 genes were significantly predictors of CRC patient’s survival.
Conclusion
Our study revealed three-gene signatures as potential therapeutic targets and also candidate molecular markers in CRC chemoradioresistance.







Similar content being viewed by others
Availability of Data
The accession number of all datasets used in this study is provided.
Code Availability
The address of all softwares used in this study was included in prepared article.
References
Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16(12):713–32.
Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14(2):89–103.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Wang X, Ghareeb WM, Lu X, Huang Y, Huang S, Chi P. Coexpression network analysis linked H2AFJ to chemoradiation resistance in colorectal cancer. J Cell Biochem. 2019;120(6):10351–62.
Sabzehali F, Azimi H, Goudarzi M. Bacteria as a vehicle in cancer therapy and drug delivery. J Paramed Sci. 2017;8(1):52–9.
Felgner S, Pawar V, Kocijancic D, Erhardt M, Weiss S. Tumour-targeting bacteria-based cancer therapies for increased specificity and improved outcome. Microb Biotechnol. 2017;10(5):1074–8.
Lee JH, Park YR, Jung M, Lim SG. Gene regulatory network analysis with drug sensitivity reveals synergistic effects of combinatory chemotherapy in gastric cancer. Sci Rep. 2020;10:3932.
Manoochehri H, Sheykhhasan M, Samadi P, Pourjafar M, Saidijam M. System biological and experimental validation of miRNAs target genes involved in colorectal cancer radiation response. Gene Rep. 2019;17: 100540.
Sun Y, Zhang Y, Wu X, Chi P. A four gene-based risk score system associated with chemoradiotherapy response and tumor recurrence in rectal cancer by co-expression network analysis. Onco Targets Ther. 2020;13:6721–33.
Luo J, Qu J, Wu D-K, Lu Z-L, Sun Y-S, Qu Q. Long non-coding RNAs: a rising biotarget in colorectal cancer. Oncotarget. 2017;8(13):22187.
Jang S-K, Yoon B-H, Kang SM, Yoon Y-G, Kim S-Y, Kim W. CDRgator: an integrative navigator of cancer drug resistance gene signatures. Cells Molecules. 2019;42(3):237–44.
Khoshinani HM, Afshar S, Pashaki AS, Mahdavinezhad A, Nikzad S, Najafi R, et al. Involvement of miR-155/FOXO3a and miR-222/PTEN in acquired radioresistance of colorectal cancer cell line. Jpn J Radiol. 2017;35(11):664–72.
Mansoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B. The different mechanisms of cancer drug resistance: a brief review. Adv Pharm Bull. 2017;7(3):339–48.
Yuan J, Tan L, Yin Z, Tao K, Wang G, Shi W, et al. Bioinformatics analysis identifies potential chemoresistance-associated genes across multiple types of cancer. Oncol Lett. 2019;18(3):2576–83.
Peng Q, Lin K, Shen Y, Zhou P, Fan S, Shen Y, et al. Identification of potential genes and pathways for response prediction of neoadjuvant chemoradiotherapy in patients with rectal cancer by systemic biological analysis. Oncol Lett. 2019;17(1):492–501.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559.
Khoei SG, Manoochehri H, Saidijam M. Systemic biological study for identification of miR-299–5p target genes in cancer. Meta Gene. 2020;24: 100655.
Lv J, Li L. Hub genes and key pathway identification in colorectal cancer based on bioinformatic analysis. Biomed Res Int. 2019;2019:1545680.
Goldstein M, Kastan MB. The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu Rev Med. 2015;66:129–43.
Jun S, Jung YS, Suh HN, Wang W, Kim MJ, Oh YS, et al. LIG4 mediates Wnt signalling-induced radioresistance. Nat Commun. 2016;7:10994.
Ribelles N, López-Siles J, Sánchez A, González E, Sánchez MJ, Carabantes F, et al. A carboxylesterase 2 gene polymorphism as predictor of capecitabine on response and time to progression. Curr Drug Metab. 2008;9(4):336–43.
Zhang G, Luo X, Zhang W, Chen E, Xu J, Wang F, et al. CXCL-13 regulates resistance to 5-Fluorouracil in colorectal cancer. Cancer Res Treat. 2020;52(2):622–33.
Storch K, Cordes N. Focal adhesion-chromatin linkage controls tumor cell resistance to radio- and chemotherapy. Chemother Res Pract. 2012;2012: 319287.
Gao J, Yan Q, Liu S, Yang X. Knockdown of EPCAM enhances the chemosensitivity of breast cancer cells to 5-fluorouracil by downregulating the antiapoptotic factor BCL-2. PLoS One. 2014;9(7): 102590.
García-Aranda M, Redondo M. Targeting receptor kinases in colorectal cancer. Cancers (Basel). 2019;11(4):433.
Wantoch von Rekowski K, König P, Henze S, Schlesinger M, Zawierucha P, Januchowski R, et al. The impact of integrin-mediated matrix adhesion on cisplatin resistance of W1 ovarian cancer cells. Biomolecules. 2019;9(12):788.
Steinbichler TB, Dudás J, Skvortsov S, Ganswindt U, Riechelmann H, Skvortsova I-I. Therapy resistance mediated by exosomes. Mol Cancer. 2019;18(1):58.
Jiang N, Dai Q, Su X, Fu J, Feng X, Peng J. Role of PI3K/AKT pathway in cancer: the framework of malignant behavior. Mol Biol Rep. 2020;47(6):4587–629.
Ciccarelli C, Vulcano F, Milazzo L, Gravina GL, Marampon F, Macioce G, et al. Key role of MEK/ERK pathway in sustaining tumorigenicity and in vitro radioresistance of embryonal rhabdomyosarcoma stem-like cell population. Mol Cancer. 2016;15(1):16.
He Z, Duan X, Zeng G. Identification of potential biomarkers and pivotal biological pathways for prostate cancer using bioinformatics analysis methods. PeerJ. 2019;7:7872.
He Y, Liu Y, Gong J, Liu C, Zhang H, Wu H. Identification of key pathways and candidate genes in pancreatic ductal adenocarcinoma using bioinformatics analysis. Oncol Lett. 2019;17(4):3751–64.
Li L, Zhu Z, Zhao Y, Zhang Q, Wu X, Miao B, et al. FN1, SPARC, and SERPINE1 are highly expressed and significantly related to a poor prognosis of gastric adenocarcinoma revealed by microarray and bioinformatics. Sci Rep. 2019;9(1):1–9.
Liu Y, Yun GH, Zhu J, Zhang C, Niu Y-M. Identification of hub genes and key pathways associated with bipolar disorder based on weighted gene co-expression network analysis. Front Physiol. 2019;10:1081.
Bonnet E, Tatari M, Joshi A, Michoel T, Marchal K, Berx G, et al. Module network inference from a cancer gene expression data set identifies microRNA regulated modules. PLoS ONE. 2010;5(4):10162.
Pawlowska E, Szczepanska J, Szatkowska M, Blasiak J. An interplay between senescence, apoptosis and autophagy in glioblastoma multiforme—role in pathogenesis and therapeutic perspective. Int J Mol Sci. 2018;19(3):889.
Manoochehri Khoshinani H, Afshar S, Najafi R. Hypoxia: a double-edged sword in cancer therapy. Cancer Invest. 2016;34(10):536–45.
Xia Y, Jiang L, Zhong T. The role of HIF-1α in chemo-/radioresistant tumors. Onco Targets Ther. 2018;11:3003–11.
Gewirtz DA. Autophagy and senescence: a partnership in search of definition. Autophagy. 2013;9(5):808–12.
Li F. Sethi G (2010) Targeting transcription factor NF-κB to overcome chemoresistance and radioresistance in cancer therapy. Biochim Biophys Acta Rev Cancer. 1805;2:167–80.
Trocoli A, Djavaheri-Mergny M. The complex interplay between autophagy and NF-κB signaling pathways in cancer cells. Am J Cancer Res. 2011;1(5):629–49.
D’Ignazio L, Rocha S. Hypoxia induced NF-κB Cells. 2016;5(1):10.
Görlach A, Bonello S. The cross-talk between NF-κB and HIF-1: further evidence for a significant liaison. Biochem J. 2008;412(3):17–9.
Sikandar SS, Pate KT, Anderson S, Dizon D, Edwards RA, Waterman ML, et al. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer Res. 2010;70(4):1469–78.
Yin L, Velazquez OC, Liu Z-J. Notch signaling: emerging molecular targets for cancer therapy. Biochem Pharmacol. 2010;80(5):690–701.
Venkatesh V, Nataraj R, Thangaraj GS, Karthikeyan M, Gnanasekaran A, Kaginelli SB, et al. Targeting Notch signalling pathway of cancer stem cells. Stem Cell Investig. 2018;5-5.
Castells M, Thibault B, Delord J-P, Couderc B. Implication of tumor microenvironment in chemoresistance: tumor-associated stromal cells protect tumor cells from cell death. Int J Mol Sci. 2012;13(8):9545–71.
Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–83.
Alimbetov D, Askarova S, Umbayev B, Davis T, Kipling D. Pharmacological targeting of cell cycle, apoptotic and cell adhesion signaling pathways implicated in chemoresistance of cancer cells. Int J Mol Sci. 2018;19(6):1690.
Yang Z, Chen H, Huo L, Yang Z, Bai Y, Fan X, et al. Epigenetic inactivation and tumor-suppressor behavior of NGFR in human colorectal cancer. Mol Cancer Res. 2015;13(1):107–19.
Lofton-Day C, Model F, DeVos T, Tetzner R, Distler J, Schuster M, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54(2):414–23.
Marchetti D, Aucoin R, Blust J, Murry B, Greiter-Wilke A. p75 neurotrophin receptor functions as a survival receptor in brain-metastatic melanoma cells. J Cell Biochem. 2004;91(1):206–15.
Jin H, Pan Y, He L, Zhai H, Li X, Zhao L, et al. p75 neurotrophin receptor inhibits invasion and metastasis of gastric cancer. Mol Cancer Res. 2007;5(5):423–33.
Passino MA, Adams RA, Sikorski SL, Akassoglou K. Regulation of hepatic stellate cell differentiation by the neurotrophin receptor p75NTR. Science. 2007;315(5820):1853–6.
Chakravarthy R, Mnich K, Gorman AM. Nerve growth factor (NGF)-mediated regulation of p75NTR expression contributes to chemotherapeutic resistance in triple negative breast cancer cells. Biochem Biophys Res Commun. 2016;478(4):1541–7.
Huang J, Yu Q, Zhou Y, Chu Y, Jiang F, Zhu X, et al. A positive feedback loop formed by NGFR and FOXP3 contributes to the resistance of non-small cell lung cancer to icotinib. Transl Cancer Res. 2020;9(2):1044–52.
Zhang Z, Fang C, Wang Y, Zhang J, Yu J, Zhang Y, et al. COL1A1: a potential therapeutic target for colorectal cancer expressing wild-type or mutant KRAS. Int J Oncol. 2018;53(5):1869–80.
Wu Y-H, Chang T-H, Huang Y-F, Chen C-C, Chou C-Y. COL11A1 confers chemoresistance on ovarian cancer cells through the activation of Akt/c/EBPβ pathway and PDK1 stabilization. Oncotarget. 2015;6(27):23748–63.
Liu S, Liao G, Li G. Regulatory effects of COL1A1 on apoptosis induced by radiation in cervical cancer cells. Cancer Cell Int. 2017;17:73.
Hosseini-Baraftabi N, Zia-Jahromi N, Talebi A. Comparison of interleukin 18 gene expression and its serum level between Iranian colorectal cancer (CRC) patients and healthy people. Biologia. 2019;74(1):103–9.
Li B, Wang F, Ma C, Hao T, Geng L, Jiang H. Predictive value of IL-18 and IL-10 in the prognosis of patients with colorectal cancer. Oncol Lett. 2019;18(1):713–9.
Yao L, Zhang Y, Chen K, Hu X, Xu LX. Discovery of IL-18 as a novel secreted protein contributing to doxorubicin resistance by comparative secretome analysis of MCF-7 and MCF-7/Dox. PLoS ONE. 2011;6(9):24684.
Korc M, Friesel RE. The role of fibroblast growth factors in tumor growth. Curr Cancer Drug Targets. 2009;9(5):639–51.
Caiado H, Conceição N, Tiago D, Marreiros A, Vicente S, Enriquez JL, et al. Data on the evaluation of FGF2 gene expression in colorectal cancer. Data Brief. 31(August 2020): 105765.
Ader I, Toulas C, Dalenc F, Delmas C, Bonnet J, Cohen-Jonathan E, et al. RhoB controls the 24 kDa FGF-2-induced radioresistance in HeLa cells by preventing post-mitotic cell death. Oncogene. 2002;21(39):5998–6006.
Kurimoto R, Iwasawa S, Ebata T, Ishiwata T, Sekine I, Tada Y, et al. Drug resistance originating from a TGF-β/FGF-2-driven epithelial-to-mesenchymal transition and its reversion in human lung adenocarcinoma cell lines harboring an EGFR mutation. Int J Oncol. 2016;48(5):1825–36.
Guo H, Chen Y, Hu X, Qian G, Ge S, Zhang J. The regulation of toll-like receptor 2 by miR-143 suppresses the invasion and migration of a subset of human colorectal carcinoma cells. Mol Cancer. 2013;12(1):1–10.
Gao F, Zhang C, Zhou C, Sun W, Liu X, Zhang P, et al. A critical role of toll-like receptor 2 (TLR2) and its’ in vivo ligands in radio-resistance. Sci Rep. 2015;5(1):1–8.
Wang Y, Liu S, Zhang Y, Yang J. Dysregulation of TLR2 serves as a prognostic biomarker in breast cancer and predicts resistance to endocrine therapy in the luminal B subtype. Front Oncol. 2020;10:547.
Li Z, Wang N, Huang C, Bao Y, Jiang Y, Zhu G. Downregulation of caveolin-1 increases the sensitivity of drug-resistant colorectal cancer HCT116 cells to 5-fluorouracil. Oncol Lett. 2017;13(1):483–7.
Park YY, An CH, Oh ST, Chang ED, Lee J. Expression of CD133 is associated with poor prognosis in stage II colorectal carcinoma. Medicine. 2019;98(32):16709.
Akbari M, Shomali N, Faraji A, Shanehbandi D, Asadi M, Mokhtarzadeh A, et al. CD133: an emerging prognostic factor and therapeutic target in colorectal cancer. Cell Biol Int. 2020;44(2):368–80.
Funding
This work was supported by the vice chancellor for Research and Technology, Hamadan University of Medical Sciences [grant number: 9811158699].
Author information
Authors and Affiliations
Contributions
Hamed Manoochehri: Data curation, conceptualization, methodology, analysis, and writing—original draft preparation. Akram Jalali: Investigation and resources. Hamid Tanzadehpanah: Writing—reviewing and editing. Amir Taherkhani: Software and editing. Massoud Saidijam: Validation, supervision, and project administration. All authors approved the paper and have substantial contributions in preparing it.
Corresponding authors
Ethics declarations
Ethics Approval
The present project was approved by the Ethics Committee of Hamadan University of Medical Sciences (Ethics Code: IR.UMSHA.REC.1398.908).
Consent to Participate
No informed consent is needed.
Consent for Publication
This article does not contain any studies with human participants.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Manoochehri, H., Jalali, A., Tanzadehpanah, H. et al. Identification of Key Gene Targets for Sensitizing Colorectal Cancer to Chemoradiation: an Integrative Network Analysis on Multiple Transcriptomics Data. J Gastrointest Canc 53, 649–668 (2022). https://doi.org/10.1007/s12029-021-00690-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-021-00690-2