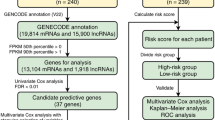
Abstract
Non-invasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP) is a low-risk thyroid tumor with a favorable prognosis. Nonetheless, differentiating NIFTP from other thyroid tumors remains challenging, necessitating reliable diagnostic markers. This study is aimed at discovering NIFTP-specific mRNA markers through RNA sequencing analysis of thyroid tumor tissues. We performed mRNA expression profiling for 74 fresh frozen thyroid tissue samples, including NIFTP and benign and malignant follicular-cell-derived tumors. NIFTP/malignant tumors showed 255 downregulated genes and 737 upregulated genes compared to benign tumors. Venn diagram analysis revealed 19 significantly upregulated and 7 downregulated mRNAs in NIFTP. Akaike information criterion analysis allowed us to select OCLN, ZNF423, LYG1, and AQP5 mRNA markers. We subsequently developed a predictive model based on logistic regression analysis using these four mRNAs, which we validated in independent samples (n = 90) using a qRT-PCR assay. This model demonstrated high accuracy in predicting NIFTP in discovery dataset (AUC (area under the receiver operating characteristic) = 0.960) and the validation dataset (AUC = 0.757). Our results suggest that OCLN, ZNF423, LYG1, and AQP5 mRNA markers might serve as reliable molecular markers for identifying NIFTP among other thyroid tumors, ultimately aiding in accurate diagnosis and management of NIFTP patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Follicular cell-derived neoplasms have three categories (i.e., benign tumors, low-risk neoplasms, and malignant neoplasms) according to the fifth edition of the World Health Organization (WHO) classification of endocrine and neuroendocrine tumors [1, 2]. Prior to 2016, non-invasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP) was classified as a subtype (non-invasive encapsulated follicular variant) of papillary thyroid carcinoma (PTC) [3, 4]. In the current WHO classification, NIFTP is categorized as a low-risk neoplasm, as it poses an extremely low risk of metastatic spread or structural recurrence [1, 2]. Although tumor cells in NIFTP have features that look like PTC, they are not malignant.
Molecular markers can provide additional information to improve the accuracy and reliability of diagnosis, especially in cases where morphological features alone might be insufficient or ambiguous. At the genomic level, NIFTP is classified as a follicular-cell derived thyroid neoplasm with RAS-like molecular profiles, whereas PTC is considered a class of BRAF-like neoplasms. However, NIFTP shows significant molecular overlaps with other follicular adenomas (FAs), invasive encapsulated follicular variant of PTC (EFVPTC), and follicular thyroid carcinoma (FTC) [1, 2, 5, 6]. These neoplasms exhibit shared molecular profiles, characterized predominantly by RAS variants and less frequently by genetic alterations in EIF1AX, EZH1, DICER1, PTEN, and TSHR genes [1, 7,8,9]. Additionally, gene fusions involving PPARG and THADA genes have been identified [1, 6, 8]. In cytologic specimens, NIFTP is often indistinguishable from benign or malignant follicular-cell derived thyroid neoplasms with RAS-like molecular profiles [9,10,11,12].
Molecular testing, which analyzes mRNA expression patterns, has been utilized to differentiate between benign and malignant tumors in cytology samples. In 2011, a 167-gene classifier utilizing mRNA expression data and machine learning (Afirma Gene Expression Classifier) was introduced for molecular testing of thyroid nodules, to identify benign nodules among indeterminate thyroid nodules detected by cytopathology [13]. A newer version of this gene expression classifier called Afirma Genomic Sequencing Classifier was introduced in 2017 [14]. This classifier employs next-generation sequencing methodology to incorporate 10,196 genes (including 1115 core genes) and machine learning to enable better distinction between benign and malignant nodules [14]. The Genomic Sequencing Classifier was trained to identify NIFTP cases as suspicious [6]. However, there is limited understanding regarding the identification of a set of differentially expressed genes that can distinguish NIFTP from other types of thyroid tumors. Protein biomarkers for diagnosing malignancy in thyroid cytology samples have also been investigated using high-throughput proteomics methods. Advanced computational and bioinformatics tools have been employed to analyze proteomics data, discern differences in protein expression between benign and malignant samples, and reveal potential protein biomarkers [15]. Nevertheless, panels employing proteomics have not been commercialized to date.
The aim of this study was to identify a set of differentially expressed genes that could accurately distinguish NIFTP from other benign and malignant follicular-cell derived thyroid neoplasms.
Materials and Methods
Study Subjects
This study received approval from the Institutional Review Board of Seoul St. Mary’s Hospital of the Catholic University of Korea (KC20TISI0766). Samples of thyroid tumor tissue and non-tumor tissue were obtained from the Biobank of Seoul St. Mary’s Hospital. Demographic and baseline characteristics of patients in the discovery and validation datasets are summarized in Table 1.
For the discovery dataset, we collected 74 fresh frozen tissue samples to profile mRNA expression levels. These samples included 10 cases of thyroid follicular nodular disease (FND), 24 cases of follicular adenoma (FA), 14 cases of non-invasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP), 6 cases of classic papillary thyroid carcinoma (PTC), 6 cases of other PTC subtypes (3 encapsulated classic PTCs with predominant follicular growth, 2 invasive encapsulated solid/trabecular PTCs, 1 diffuse sclerosing PTC), and 14 cases of follicular thyroid carcinoma (FTC).
The validation dataset comprised a total of 90 fresh frozen thyroid samples, including normal thyroid tissue (n = 12), FND (n = 13), FA (n = 18), NIFTP (n = 10), FTC (n = 10), invasive encapsulated follicular variant of papillary thyroid carcinoma (IEFVPTC, n = 14), and PTC (n = 13). The specific histologic subtypes are described in Table 1. These tissue samples were obtained from different years compared to the samples in the discovery dataset.
All tumor samples utilized in this study were evaluated for the status of the BRAF V600E variant using real-time PCR PNA clamping technology (Panagene, Daejeon, Korea) [16]. All samples in the discovery dataset were deliberately selected to be negative for the BRAF V600E variant. This intentional inclusion of BRAF V600E negative tumors in the discovery dataset aimed to exclude any gene expression changes induced by the BRAF V600E. This distinction is particularly relevant since NIFTP and other follicular-patterned tumors (FND, FA, and FTC) do not possess the BRAF V600E mutation. In contrast, the validation dataset included 11 PTCs with the BRAF V600E variant. This decision was made because the selected mRNA markers needed to be effective in differentiating NIFTP from other follicular cell-derived neoplasms, including PTCs with BRAF V600E.
Total RNA Preparation and mRNA Sequencing
Total RNAs were isolated from fresh-frozen tissues using a RNeasy Kit (Qiagen, Carlsbad, CA, USA) following the manufacturer’s instructions. The quantity and quality of obtained total RNAs were evaluated using an ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). RNA integrity number (RIN) was estimated using a 2100 Agilent Bioanalyzer (Agilent Technologies, Waldbronn, Germany). Approximately 1 μg of total RNAs was utilized for library preparation with an Illumina TruSeq Stranded Total RNA Library Prep Kit (San Diego, CA, USA). An Illumina NovaSeq 6000 System sequencing instrument was employed to perform 101-bp paired-end sequencing, generating following the manufacturer’s instructions.
mRNA Sequencing Data Analysis in the Discovery Dataset
We eliminated TruSeq small RNA adapters from sequenced reads using Trimmomatic software (v. 0.38). Remaining sequence data were mapped to the human genome (GRCh38) for quantification using STAR software (v. 2.7.a) [17]. To identify differentially expressed mRNA, the edgeR software was employed [18]. We utilized the default parameter configuration for all programs. The mRNA-seq dataset is available from Korean Nucleotide Archive (KoNA, https://kobic.re.kr/kona) and Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) public databases (accession number: PRJKA220514 and PRJNA918826, respectively).
mRNA Expression Levels by qRT-PCR in the Validation Dataset
In the validation dataset, mRNA expression levels were evaluated using quantitative reverse-transcription PCR (qRT-PCR). To assess the expression levels of selected candidate mRNAs for the identification of NIFTP, 2 µg of total RNA from each specimen was converted into cDNA using the iScript reverse transcriptase kit (Bio-Rad, Hercules, CA). The subsequent real-time RT-PCR procedure was performed using the iQ SYBR Green Supermix (Bio-Rad) on a CFX96 real-time PCR system (Bio-Rad). The quantified mRNA levels were normalized to β-actin levels. Detailed information about primer sequences and the experimental conditions used are provided in Table S1.
Public mRNA Sequencing Data Analysis
We obtained public mRNA-sequencing data of thyroid samples from The Cancer Genome Atlas (TCGA) dataset (https://portal.gdc.cancer.gov/) to confirm mRNA expression patterns of candidate mRNA markers.
Statistical Analysis
To evaluate significance of difference in gene expression between benign and NIFTP/malignant tumor tissues, Student’s t-test or analysis of variance (ANOVA) was used. We performed hierarchical clustering using Multiple Experiment Viewer (MEV) software (version 4.8.1) and Pearson’s correlation method [19]. For Kyoto Encyclopedia of Genes and Genomes (KEGG) Gene Ontology (GO) enrichment analysis, we used the ShinyGO tool [20]. Using ROCR package of R software (version 4.2.2), we calculated receiver operating characteristic (ROC) and area under the ROC curve (AUC) for each mRNA marker. We performed logistic regression for the combination of mRNA markers to identify NIFTP from other thyroid tumors. The prediction score was calculated by multiplying expression level of each mRNA with its corresponding regression coefficient and then summing them up in a linear combination. We estimated optimal cutoff values maximizing sensitivity and specificity between low and high levels of mRNA expression using ROC curve analysis. Results with p values < 0.05 were considered significant.
Results
Global Differential Gene Expression Between Benign and NIFTP/Malignant Thyroid Tumors
In this study, we conducted mRNA expression profiling of 74 fresh frozen thyroid tissues, including thyroid benign (FND and FA), NIFTP, and malignant (PTC and FTC) tumors. Notably, we only included PTCs that lacked BRAF V600E variant. To identify differentially expressed genes (DEGs), we applied two criteria: (1) p value < 0.05 and (2) log2 fold change > 0.5 between benign thyroid tumors (FA and FND) and NIFTP/malignancy (FTC and PTC). Based on these criteria, we identified 255 downregulated and 737 upregulated genes in NIFTP/malignancy compared to benign tumors. Figure 1A shows results of unsupervised hierarchical clustering using these DEG candidates.
mRNA expression profiling for fresh frozen thyroid tumor tissues (n = 74). A Unsupervised hierarchical cluster analysis was conducted to examine the expression levels of 255 downregulated and 737 upregulated genes in NIFTP and malignancy, which were identified using the edgeR software. RNA expression levels were normalized by Z-score. B KEGG pathway enrichment analysis was performed for differentially expressed genes (DEGs) between benign and NIFTP/malignant thyroid tumors
Next, we performed KEGG pathway enrichment analysis using DEGs between benign tumors and NIFTP/malignancy and found several cancer-associated pathways, including ECM-receptor interaction, MAPK signaling pathway, Ras signaling pathway, pathway in cancer, and p53 signaling pathway (Fig. 1B). Our results demonstrate that RNA-seq platform can successfully identify DEGs between benign and NIFTP/malignant thyroid tumors.
Identification of Differentially Expressed mRNAs in NIFTP
We analyzed differentially expressed mRNAs between various tumor subgroups (FND vs. NIFTP, FA vs. NIFTP, FTC vs. NIFTP, and PTC vs. NIFTP) to identify mRNA markers exclusive to NIFTP. Using Venn diagrams, we identified 19 significantly upregulated mRNAs and 7 significantly downregulated mRNAs in NIFTP (Fig. 2A). We performed unsupervised hierarchical clustering of candidate mRNA markers. Results are shown in Fig. 2B.
Identification of NIFTP-specific mRNA markers in the discovery dataset. A Comparative analysis of differentially expressed mRNAs between NIFTP and other subtypes (FND, FA, FTC, and PTC) revealed 19 upregulated and 7 downregulated mRNAs in NIFTP. B A heatmap of differentially expressed mRNAs was generated for the thyroid tumor subgroup. RNA expression levels were normalized to Z-score
To further refine our list of NIFTP-specific candidate mRNA markers, we analyzed the TCGA dataset. The TCGA project did not enroll cases of NIFTP since the tumor was not yet recognized as a distinct entity until after the TCGA project was completed in 2016 [3, 21]. In this study, we reviewed whole slide images available in the TCGA dataset and observed that encapsulated follicular subtype of PTC might encompass cases that could potentially be classified as NIFTPs. We excluded follicular PTCs with BRAF V600E-like molecular alterations. Although some of the encapsulated follicular PTCs might not meet the criteria for NIFTP diagnosis, they were nevertheless instrumental in validating our results. Out of the 26 candidate mRNAs identified in our own samples, 12 of them exhibited distinct expression levels between the follicular subtype and other subtypes of PTC in the TCGA dataset.
Out of the 12 selected mRNAs, 8 (KCNC1, PDE1B, NXPH4, ZNF423, MAPT, SRMS, OCLN, and IGF2BP3) were upregulated and 4 (AQP5, LYG1, IDO1, and PHYHD1) were downregulated in NIFTP as shown in Table 2. These 12 mRNAs were incorporated into subsequent downstream analyses.
Combination of mRNA Markers to Identify NIFTP
To determine the optimal combination of mRNA markers for identifying NIFTP, we conducted an Akaike information criterion (AIC) analysis using the top 12 candidate mRNA markers. We ultimately selected OCLN (occludin), ZNF423 (zinc finger protein 423), LYG1 (lysozyme g1), and AQP5 (aquaporin 5) mRNA markers based on their expression pattern in the discovery dataset (Fig. 3).
Multiple mRNA marker selection for identification of non-invasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP) in the discovery dataset. A Akaike information criterion (AIC) analysis was performed using 12 candidate mRNA markers to discriminate NIFTP with Blorr R software packages. Genes selected by the AIC included OCLN, ZNF423, LYG1, and AQP5. B Expression patterns of the 12 selected four mRNA markers in each thyroid tumor were analyzed using RNA expression levels normalized to log2 CPM. ANOVA analysis was used to estimate statistical significance
To estimate the accuracy of these four NIFTP-specific mRNA markers, we performed ROC analysis and calculated AUC values. AUC values for OCLN, ZNF423, LYG1, and AQP5 were 0.80, 0.73, 0.73, and 0.68, respectively (Table 3).
We then developed the following equation for predicting NIFTP using a logistic regression analysis of the four mRNAs: prediction score = −25.120 + (3.797 × expression level of OCLN) + (3.690 × expression level of ZNF423) + (−5.229 × expression level of LYG1) + (−0.756 × expression level of AQP5). The prediction model had an AUC value of 0.960 in the discovery dataset (Fig. 4A).
Validation of mRNA Markers to Identify NIFTP in an Independent Cohort
The four NIFTP-specific mRNA markers were validated in independent samples using a qRT-PCR assay. These samples encompassed normal thyroid tissue, FND, FA, NIFTP, FTC, IEFVPTC, and PTC. All four mRNA markers demonstrated significant differential expression across these tumor types (Fig. 5). We then generated a prediction equation for NIFTP using a logistic regression analysis of these four mRNAs to assess their capacity to distinguish NIFTP from other tumor types: prediction score = −3.722 + (9.950 × expression level of OCLN) + (70.667 × expression level of ZNF423) - (273.325 × expression level of LYG1) + (2.752 × expression level of AQP5). The prediction model demonstrated a promising AUC value in the validation dataset (AUC = 0.757, Fig. 4B). The performance of the model is summarized in Table 4.
Expression levels of selected four mRNA markers in validation dataset using qRT-PCR. Expression levels of OCLN (A), ZNF423 (B), LYG1 (C), and AQP5 (D) are shown in relation to normal thyroid tissue and various tumor types. FND, thyroid follicular nodular disease; FA, follicular adenoma; NIFTP, non-invasive follicular thyroid neoplasm with papillary-like nuclear features; IEFVPTC, invasive encapsulated follicular variant of papillary thyroid carcinoma; FTC, follicular thyroid carcinoma; PTC, papillary thyroid carcinoma
Our findings suggest that these four mRNA markers can serve as reliable molecular markers for identifying NIFTP among other thyroid tumors.
Discussion
We identified 19 significantly upregulated mRNAs and 7 significantly downregulated mRNAs in NIFTP compared to various other tumor subgroups in the discovery cohort. Further analyses using TCGA dataset and AIC method led us to select OCLN, ZNF423, LYG1, and AQP5 mRNA markers as optimal markers for identifying NIFTP. In addition, ROC analysis showed good accuracy for predicting NIFTP using these four mRNA markers in discovery and validation datasets. Our results suggest that these four mRNA markers can serve as reliable molecular markers for identifying NIFTP among other thyroid tumors.
Developing mRNA markers for NIFTP could potentially help differentiate it from other follicular cell-derived thyroid tumors with overlapping morphological features, which can have different clinical management and prognostic implications. These mRNA markers might help reduce overtreatment of indolent tumors and guide more appropriate therapeutic interventions for patients with thyroid cancer. Identification of specific mRNA markers might provide insights into the underlying biology of NIFTP and help further our understanding of its pathogenesis. Additionally, investigating functional roles of these candidate markers in thyroid tumor development and progression could provide insights into molecular mechanisms underlying NIFTP and might reveal potential therapeutic targets for treating thyroid tumors.
In NIFTP, of the four mRNAs investigated, two (OCLN and ZNF423) exhibited upregulation while the other two (LYG1 and AQP5) displayed downregulation. To date, no studies have examined the roles of these genes in the development or progression of thyroid tumors. Nonetheless, in other cancer types, these genes have been implicated in functioning as either oncogenes or tumor suppressors, depending on the specific tumor context. OCLN encodes a protein called occludin, which is an integral membrane protein and a key component of tight junctions between cells [22]. Tight junctions are essential for maintaining cell polarity and regulating the passage of molecules between cells. Downregulation of OCLN has been observed in clear cell renal cell carcinoma [23], hepatocellular carcinoma [24], breast cancer [25], endometrial cancer, and lung cancer [26]. However, OCLN is overexpressed in bladder cancer. It regulates angiogenesis in bladder cancer [27]. ZNF423 is a transcription factor that plays a role in regulating the expression of various target genes involved in development, cell differentiation, and DNA damage response [28]. ZNF423 has been implicated in the regulation of tumor growth, cell proliferation, and apoptosis [29]. Altered ZNF423 expression has been reported in different cancers, including breast cancer [30], ovary cancer [30], and neuroblastoma [31]. Its role in cancer progression might be context-dependent, acting as either an oncogene or tumor suppressor depending on the specific tumor type and cellular context [32]. LYG1 belongs to lysozyme G family. Its function in mammalian cells and its role in cancer are not well understood yet. However, LYG1 may play a role in antitumor function by promoting the activation, proliferation, and function of CD4 + T cells in tumor microenvironment [33]. AQP5 encodes a water channel protein that is responsible for facilitating water transport across cell membranes. Aquaporins, including AQP5, have been implicated in various aspects of cancer biology, such as cell migration, proliferation, and angiogenesis [34,35,36]. Altered AQP5 expression has been observed in multiple cancer types, including lung cancer [34], breast cancer [35], and colorectal cancer [36]. Overexpression of AQP5 has been associated with increased tumor growth, metastasis, and poor prognosis in some cancer types [37], while its downregulation has been linked to reduced tumor growth and metastasis in non-small cell lung cancer cells [38].
This study is aimed at identifying NIFTP-specific candidate mRNA markers by analyzing both our own datasets and the publicly available TCGA dataset. While the TCGA project did not enroll cases of NIFTP due to its recognition as a distinct entity after the project’s completion, we were able to leverage the TCGA dataset by reviewing whole slide images and selecting encapsulated follicular subtype of PTCs that could potentially be classified as NIFTPs. This approach allowed us to refine our list of candidate mRNA markers. It is important to acknowledge that some of the identified cases in the TCGA dataset might not meet the strict criteria for NIFTP diagnosis. Given that this limitation could potentially affect the specificity of our findings, we primarily utilized the TCGA dataset to narrow down the list of mRNA markers. The diagnostic performance of the final four selected markers was then validated within an independent cohort from our study. This supports the validity of our results and highlights the potential of these markers for differentiating NIFTP from other thyroid tumors.
In this study, we observed the remarkable diagnostic performance of four mRNA markers within the discovery dataset. However, when these findings were subjected to validation in an independent cohort, there was a decrease in diagnostic performance. Several factors could contribute to this disparity in results. First, it is common to observe optimized results within the discovery dataset because the markers were derived and tested within the same dataset. However, the validation dataset introduces real-world scenarios, which could lead to biological variability and possibly different pre-analytical and analytical conditions, thereby influencing the markers’ performance. Second, there were differences in the methodologies employed for analyzing mRNA expression levels between datasets. Specifically, next-generation sequencing was used in the discovery dataset, while qRT-PCR was employed for the validation dataset. Third, it is plausible that the selected mRNA markers are not as universally applicable as initially anticipated. While they demonstrated impressive performance under the controlled conditions of the discovery dataset, their applicability might have been overestimated when considering a broader, more diverse population, as represented in the validation dataset. This suggests that there may be a need to identify additional markers to enhance diagnostic accuracy across varied cohorts. The decreased performance in the validation dataset does not invalidate our findings but suggests that additional refinement and validation of the markers are necessary. Future studies should focus on further improving the diagnostic accuracy of these markers, combining them into a multi-marker panel, or integrating them with clinicopathological parameters in a diagnostic algorithm. Despite these limitations, our study represents an important step forward in the search for reliable, non-invasive diagnostic markers for NIFTP. Our findings provide a foundation upon which further research can build, bringing us closer to the goal of a more accurate diagnosis of low-risk thyroid neoplasms.
Conclusion
Our study provides evidence that OCLN, ZNF423, LYG1, and AQP5 mRNA markers can serve as reliable molecular markers for identifying NIFTP among other thyroid tumors. Further research is needed to validate these findings in larger patient cohorts and explore molecular mechanisms underlying the roles of these markers in NIFTP development. Ultimately, implementation of these markers in clinical practice could improve diagnostic accuracy and inform more tailored treatment strategies for patients with thyroid tumors.
Availability of Data and Material
The mRNA-seq dataset is available in the Korean Nucleotide Archive (KoNA, https://kobic.re.kr/kona) and Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) public databases under accession numbers PRJKA220514 and PRJNA918826.
Code Availability
Not applicable.
References
Baloch ZW, Asa SL, Barletta JA, Ghossein RA, Juhlin CC, Jung CK, et al. Overview of the 2022 WHO Classification of Thyroid Neoplasms. Endocr Pathol. 2022;33(1):27-63. https://doi.org/10.1007/s12022-022-09707-3.
Jung CK, Bychkov A, Kakudo K. Update from the 2022 World Health Organization Classification of Thyroid Tumors: A Standardized Diagnostic Approach. Endocrinol Metab (Seoul). 2022;37(5):703-18. https://doi.org/10.3803/EnM.2022.1553.
Nikiforov YE, Seethala RR, Tallini G, Baloch ZW, Basolo F, Thompson LD, et al. Nomenclature Revision for Encapsulated Follicular Variant of Papillary Thyroid Carcinoma: A Paradigm Shift to Reduce Overtreatment of Indolent Tumors. JAMA Oncol. 2016;2(8):1023-9. https://doi.org/10.1001/jamaoncol.2016.0386.
Katsakhyan L, Song S, Lepe M, Shojaei H, Montone KT, LiVolsi VA, et al. Practice Paradigms Before and After Introduction of the Diagnosis-Noninvasive Follicular Thyroid Neoplasm with Papillary-Like Nuclear Features (NIFTP): an Institutional Experience. Endocr Pathol. 2020;31(2):174-81. https://doi.org/10.1007/s12022-020-09614-5.
Brandler TC, Zhou F, Liu CZ, Serrano A, Sun W, Nikiforov YE, et al. Molecular Profiles of Non-Invasive, Minimally Invasive and Invasive Follicular Patterned Thyroid Neoplasms with Papillary Nuclear Features. Thyroid. 2023. https://doi.org/10.1089/thy.2023.0047.
Rossi ED, Faquin WC, Baloch Z, Fadda G, Thompson L, Larocca LM, et al. Noninvasive Follicular Thyroid Neoplasm with Papillary-Like Nuclear Features (NIFTP): Update and Diagnostic Considerations-a Review. Endocr Pathol. 2019;30(2):155-62. https://doi.org/10.1007/s12022-019-9574-7.
Bae JS, Jung SH, Hirokawa M, Bychkov A, Miyauchi A, Lee S, et al. High Prevalence of DICER1 Mutations and Low Frequency of Gene Fusions in Pediatric Follicular-Patterned Tumors of the Thyroid. Endocr Pathol. 2021;32(3):336-46. https://doi.org/10.1007/s12022-021-09688-9.
Agarwal S, Bychkov A, Jung CK. Emerging Biomarkers in Thyroid Practice and Research. Cancers (Basel). 2021;14(1). https://doi.org/10.3390/cancers14010204.
Chin PD, Zhu CY, Sajed DP, Fishbein GA, Yeh MW, Leung AM, et al. Correlation of ThyroSeq Results with Surgical Histopathology in Cytologically Indeterminate Thyroid Nodules. Endocr Pathol. 2020;31(4):377-84. https://doi.org/10.1007/s12022-020-09641-2.
Bongiovanni M, Giovanella L, Romanelli F, Trimboli P. Cytological Diagnoses Associated with Noninvasive Follicular Thyroid Neoplasms with Papillary-Like Nuclear Features According to the Bethesda System for Reporting Thyroid Cytopathology: A Systematic Review and Meta-Analysis. Thyroid. 2019;29(2):222-8. https://doi.org/10.1089/thy.2018.0394.
Na HY, Park SY. Noninvasive follicular thyroid neoplasm with papillary-like nuclear features: its updated diagnostic criteria, preoperative cytologic diagnoses and impact on the risk of malignancy. J Pathol Transl Med. 2022;56(6):319-25. https://doi.org/10.4132/jptm.2022.09.29.
Chen CC, Hang JF, Liu CY, Wang YH, Lai CR. Thyroid fine-needle aspiration cytology in Taiwan: a nationwide survey and literature update. J Pathol Transl Med. 2020;54(5):361-6. https://doi.org/10.4132/jptm.2020.07.17.
Alexander EK, Kennedy GC, Baloch ZW, Cibas ES, Chudova D, Diggans J, et al. Preoperative Diagnosis of Benign Thyroid Nodules with Indeterminate Cytology. N Engl J Med. 2012;367(8):705-15. https://doi.org/10.1056/nejmoa1203208.
Patel KN, Angell TE, Babiarz J, Barth NM, Blevins T, Duh QY, et al. Performance of a Genomic Sequencing Classifier for the Preoperative Diagnosis of Cytologically Indeterminate Thyroid Nodules. JAMA Surg. 2018;153(9):817-24. https://doi.org/10.1001/jamasurg.2018.1153.
Nikas IP, Ryu HS. The application of high-throughput proteomics in cytopathology. J Pathol Transl Med. 2022;56(6):309-18. https://doi.org/10.4132/jptm.2022.08.30.
Kim SY, Kim T, Kim K, Bae JS, Kim JS, Jung CK. Highly prevalent BRAF V600E and low-frequency TERT promoter mutations underlie papillary thyroid carcinoma in Koreans. J Pathol Transl Med. 2020;54(4):310-7. https://doi.org/10.4132/jptm.2020.05.12.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21. https://doi.org/10.1093/bioinformatics/bts635.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139-40. https://doi.org/10.1093/bioinformatics/btp616.
Howe EA, Sinha R, Schlauch D, Quackenbush J. RNA-Seq analysis in MeV. Bioinformatics. 2011;27(22):3209-10. https://doi.org/10.1093/bioinformatics/btr490.
Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36(8):2628-9. https://doi.org/10.1093/bioinformatics/btz931.
Cancer Genome Atlas Research N. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676–90. https://doi.org/10.1016/j.cell.2014.09.050.
Bendriem RM, Singh S, Aleem AA, Antonetti DA, Ross ME. Tight junction protein occludin regulates progenitor Self-Renewal and survival in developing cortex. Elife. 2019;8. https://doi.org/10.7554/eLife.49376.
Conceição ALG, Da Silva CT, Badial RM, Valsechi MC, Stuqui B, Gonçalves JD, et al. Downregulation of OCLN and GAS1 in clear cell renal cell carcinoma. Oncol Rep. 2017;37(3):1487-96. https://doi.org/10.3892/or.2017.5414.
Orban E, Szabo E, Lotz G, Kupcsulik P, Paska C, Schaff Z, et al. Different expression of occludin and ZO-1 in primary and metastatic liver tumors. Pathol Oncol Res. 2008;14(3):299-306. https://doi.org/10.1007/s12253-008-9031-2.
Martin TA, Mansel RE, Jiang WG. Loss of occludin leads to the progression of human breast cancer. Int J Mol Med. 2010;26(5):723-34. https://doi.org/10.3892/ijmm_00000519.
Tobioka H, Isomura H, Kokai Y, Tokunaga Y, Yamaguchi J, Sawada N. Occludin expression decreases with the progression of human endometrial carcinoma. Hum Pathol. 2004;35(2):159-64. https://doi.org/10.1016/j.humpath.2003.09.013.
Yang F, Liu XQ, He JZ, Xian SP, Yang PF, Mai ZY, et al. Occludin facilitates tumour angiogenesis in bladder cancer by regulating IL8/STAT3 through STAT4. J Cell Mol Med. 2022;26(8):2363-76. https://doi.org/10.1111/jcmm.17257.
Casoni F, Croci L, Bosone C, D'Ambrosio R, Badaloni A, Gaudesi D, et al. Zfp423/ZNF423 regulates cell cycle progression, the mode of cell division and the DNA-damage response in Purkinje neuron progenitors. Development. 2017;144(20):3686-97. https://doi.org/10.1242/dev.155077.
Harder L, Puller AC, Horstmann MA. ZNF423: Transcriptional modulation in development and cancer. Mol Cell Oncol. 2014;1(3):e969655. https://doi.org/10.4161/23723548.2014.969655.
Bond HM, Scicchitano S, Chiarella E, Amodio N, Lucchino V, Aloisio A, et al. ZNF423: A New Player in Estrogen Receptor-Positive Breast Cancer. Front Endocrinol (Lausanne). 2018;9:255. https://doi.org/10.3389/fendo.2018.00255.
Huang S, Laoukili J, Epping MT, Koster J, Hölzel M, Westerman BA, et al. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell. 2009;15(4):328-40. https://doi.org/10.1016/j.ccr.2009.02.023.
Chaiprasert T, Armartmuntree N, Techasen A, Sakonsinsiri C, Pinlaor S, Ungarreevittaya P, et al. Roles of Zinc Finger Protein 423 in Proliferation and Invasion of Cholangiocarcinoma through Oxidative Stress. Biomolecules. 2019;9(7). https://doi.org/10.3390/biom9070263.
Liu H, Zhang Y, Liu Z, Wang P, Mo X, Fu W, et al. LYG1 exerts antitumor function through promoting the activation, proliferation, and function of CD4(+) T cells. Oncoimmunology. 2017;6(4):e1292195. https://doi.org/10.1080/2162402X.2017.1292195.
Zhang Z, Chen Z, Song Y, Zhang P, Hu J, Bai C. Expression of aquaporin 5 increases proliferation and metastasis potential of lung cancer. J Pathol. 2010;221(2):210-20. https://doi.org/10.1002/path.2702.
Jung HJ, Park JY, Jeon HS, Kwon TH. Aquaporin-5: a marker protein for proliferation and migration of human breast cancer cells. PLoS One. 2011;6(12):e28492. https://doi.org/10.1371/journal.pone.0028492.
Wang W, Li Q, Yang T, Bai G, Li D, Li Q, et al. Expression of AQP5 and AQP8 in human colorectal carcinoma and their clinical significance. World J Surg Oncol. 2012;10:242. https://doi.org/10.1186/1477-7819-10-242.
Woo J, Lee J, Chae YK, Kim MS, Baek JH, Park JC, et al. Overexpression of AQP5, a putative oncogene, promotes cell growth and transformation. Cancer Lett. 2008;264(1):54-62. https://doi.org/10.1016/j.canlet.2008.01.029.
Elkhider A, Wang B, Ouyang X, Al-Azab M, Walana W, Sun X, et al. Aquaporin 5 promotes tumor migration and angiogenesis in non-small cell lung cancer cell line H1299. Oncol Lett. 2020;19(3):1665-72. https://doi.org/10.3892/ol.2020.11251.
Funding
This research was supported by “Systemic Industrial Infrastructure Projects” through the Ministry of Trade, Industry and Energy (MOTIE) (P0009796, 2019) and the KRIBB Research Initiative Program (KGM5192322). This research was also supported by a grant (HI21C0940) from the Korean Health Technology R&D Project funded by the Ministry of Health and Welfare, and a grant (NRF-2020R1F1A1070028) from the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science and ICT, Republic of Korea. This study was supported by a grant (NRF-2020M3A9I6A01036057) from the National Research Foundation funded by the Korea government.
Author information
Authors and Affiliations
Contributions
Conceptualization: C.K.J. and S.Y.K.; sample and data curation: S.J.Y., J.L.P., K.K., J.S.B., and J.W.K.; RNA-seq data generation and analysis: S.Y.L., J.W.K, and J.L.P.; writing—original draft: S.Y.L., J.L.P., and C.K.J.; writing—review and editing: S.Y.L., J.L.P., K.K., J.S.B, S.Y.K., and C.K.J.; supervision: C.K.J. and S.Y.K. All authors have read and agreed to the submitted version.
Corresponding authors
Ethics declarations
Ethics Approval
This study was conducted according to the guidelines of the Declaration of Helsinki. It was approved by the Institutional Review Board of Seoul St. Mary’s Hospital of the Catholic University of Korea (KC20TISI0766).
Consent for Publication
All authors consent to the publication of this paper.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, SY., Park, JL., Kim, K. et al. Identification of NIFTP-Specific mRNA Markers for Reliable Molecular Diagnosis of Thyroid Tumors. Endocr Pathol 34, 311–322 (2023). https://doi.org/10.1007/s12022-023-09781-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12022-023-09781-1