Abstract
This study aimed to evaluate how excess selenium induces oxidative stress by determining antioxidant enzyme activity and changes in expression of selected selenoproteins in mice. BALB/c mice (n = 20 per group) were fed a diet containing 0.045 (Se-marginal), 0.1 (Se-adequate), 0.4 (Se-supernutrition), or 0.8 (Se-excess) mg Se/kg. Gene expression was quantified in RNA samples extracted from the liver, kidney, and testis by real-time quantitative reverse transcription-polymerase chain reaction. We found that glutathione peroxidase (GPx) and catalase activities decreased in livers of mice fed the marginal or excess dose of Se as compared to those in the Se-adequate group. Additionally, superoxide dismutase and glutathione reductase activities were significantly reduced only in mice fed the excess Se diet, compared to animals on the adequate Se diet. Se-supernutrition had no effect on hepatic mRNA levels of GPx isoforms 1 and 4 (GPx1 and GPx4), down-regulated GPx isoform 3 (GPx3), and upregulated selenoprotein W (SelW) mRNA expression. The excess Se diet led to decreased hepatic mRNA levels of GPx1, GPx3 and GPx4 but no change in testicular mRNA levels of GPx1, GPx3 or SelW. Dietary Se had no effect on testicular mRNA levels of GPx4. Thus, our results suggest that Se exposure can reduce hepatic antioxidant capacity and cause liver dysfunction. Dietary Se was found to differentially regulate mRNA levels of the GPx family or SelW, depending on exposure. Therefore, these genes may play a role in the toxicity associated with Se.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Selenium (Se) is an essential nutritional trace element, which is best known for its antioxidant properties that are mediated by various selenoproteins. At least 25 selenoproteins in humans and 24 homologues in rodents have been identified [1]. They play important roles in diverse physiological processes such as chemoprevention, neurobiology, aging, immunity (i.e., immune responses, anti-inflammatory activity and antiviral activity), muscle metabolism, reproduction, and redox reactions [2]. The synthesis of selenoproteins is affected by levels of Se supplementation. Broadly, the field of biological Se research can be divided into two distinct areas: study of the Se nutritional essentiality and study of Se toxicity [3].
Both excessive and insufficient Se intake can result in adverse health effects [4–7]. There are numerous reports regarding the effects of Se deficiency. Gastrointestinal glutathione peroxidase (GI-GPx/GPx2) mRNA increases in Se-deficient mice [8], whereas GPx isoform 4 (GPx4) mRNA decreases substantially in Se-deficient turkey liver [9]. The levels of GPx isoform 1 (GPx1), selenoprotein W (SelW), and selenoprotein H (SelH) are dramatically decreased in Se-deficient, compared to Se-replete, rats [10]. Severe Se deficiency causes almost total loss of GPx1 activity and mRNA in the rat liver and heart, while GPx4 activity is reduced by 75 % in the liver and 60 % in the heart, leaving mRNA levels unchanged [6]. Liver Se concentration and liver GPx1 and thioredoxin reductase (Txnrd) activities were reduced in Se-deficient mice compared to Se-adequate mice [11].
Several groups have also examined the effects of excess Se intake. At high concentrations, selenite is known to induce apoptosis in cells by a mechanism involving free radicals [12]. Further, Se and GPx levels are increased in animals treated with an excess of Se [13]. GPx mRNA levels and activities were significantly increased in rats injected with 20 mg Se/kg per day, but dramatically decreased upon injection of 40 or 80 mg Se/kg per day [4]. GPx4 mRNA level in chicken was down-regulated by an excess of Se [14]. When dietary Se is increased from 0.3 to 3.0 mg Se/kg, testicular mRNA levels of Txnrd1 and selenoprotein 15 (Sep15) are attenuated, whereas expression of Gpx1 is increased in the pig liver [15]. A significant decrease in total superoxide dismutase (SOD) activity has been reported in animals with both excess and depleted Se [16].
The above results show that Se supplementation in animals has been extensively studied. There is evidence to suggest that excess selenite and selenomethionine [17, 18] can impair the amount and/or activity of biological antioxidant defense mechanisms by generation of oxygen-free radicals [19]. The Nutritional Prevention of Cancer Trial found that Se supplementation increased the risk of squamous cell carcinoma, total cancer incidence, and diabetes in subjects with higher plasma Se levels [20].
Despite this, little information is available on the effects of Se overexposure on GPx and SelW1 gene expression in mice. Examining the modulation of selenoprotein expression upon Se exposure may help elucidate the molecular mechanisms underlying Se toxicity. The aim of this study, therefore, was to analyze the effects of varying the levels of dietary exposure to Se on antioxidant capacity, as measured by GPx and SelW gene expression, and evaluate the dose-dependent effect of dietary supplementation containing a range of Se levels on regulation of redox status in different mouse tissues.
Materials and Methods
Animals and diets
All experimental protocols were approved by the ethics committee of South China Agricultural University. A total of eighty 7-week-old male BALB/c mice (Certification no. L3623, Medical Experimental Animal Center of Guangdong Province, Guangzhou) with an average body weight (BW) of 20 g were assigned to four groups, each of which was replicated twice with ten mice per replicate. Mice in Group 1 were fed a basal diet containing 0.045 mg Se/kg, while mice in Groups 2, 3 and 4 were fed the basal diet supplemented with 0.055, 0.355, and 0.755 mg Se/kg, respectively. Thus, the final concentrations of Se used in this work were 0.045, 0.1, 0.4, and 0.8 mg/kg. The basal diet was calculated to be adequate in protein, energy, vitamins, and minerals for this class of animal, except for Se content (0.045 mg/kg). The total test period was 56 days. All mice were fed with the same basal diet for 7 days to adjust the Se status and to adapt to the new environment before the experimental period.
Excretion and tissue sampling
Mice were housed in metabolic cages. Ten mice from each treatment group were selected, fasted for 12 h, and then tissues and fluids were harvested at the end of the experiment under general halothane anesthesia. All blood samples were collected in sterile tubes to assay serum biochemical parameters as described below. Samples of liver, kidney and testes were excised from individual mice, weighed immediately in an aseptic environment, then frozen in liquid nitrogen and stored at −80 °C until analysis. In this process, segmental samples of liver were weighed, homogenized on ice using a glass-Teflon homogenizer (K5424, Glas-col; Terre Haute, IN, USA), and centrifuged at 3,000×g for 10 min at 4 °C. Supernatants were used immediately for further biochemical assays, and protein concentrations were determined with the Bradford assay [21] using bovine serum albumin as a protein standard.
Determination of biochemical parameters
Biochemical parameters, including serum aspartate aminotransferase (AST), alanine aminotransferase (ALT), blood urea nitrogen (BUN) and serum creatinine (SCr), were determined with the CX-4 Auto-Blood Biochemical Analyzer (Beckman Coulter Inc.; Brea, CA, USA). Activities of redox enzymes (GPx, SOD, glutathione reductase (GR), catalase (CAT), and levels of malondialdehyde (MDA) in the liver were determined using reagent kits from the Nanjing Jiancheng Bioengineering Institute (Nanjing, China). The detection range of the kits was 0.2–1000 U/mL, the lower limit of detection was less than 0.1 U/mL, and the inter-assay and intra-assay variation coefficients were 8 and 5 %, respectively. All samples were tested in duplicate.
Assay of Se content in tissues
Tissue samples were digested with 5:1 HNO3/H2O2 in separate series. The digestions were carried out according to the program given in Table 1. The microwave digests were cooled to room temperature, which were accelerated by immersing the segments (vial holders) into an icy water bath. Volumes of the digests and reagent blanks were then adjusted to 10.0 mL with ultrapure water. The working solution was prepared in 3.0 M HCl and Se was directly measured with hydride generation-atomic fluorescence spectrometry (AFS-9130, Beijing Titan Instruments Co., Ltd.; Beijing, China). The primary standard Se (IV) solution of 1000 mg/L was prepared in 3.0 M HCl from SeO2 (99.8 %; Aldrich Chemical Co.; Milwaukee, WI, USA) and stored in a refrigerator at 4 °C. This solution is stable for at least 12 months.
Determination of selenoprotein mRNA levels in tissues
Total RNA was isolated from tissue samples using Trizol reagent (Invitrogen; Carlsbad, CA, USA), according to the manufacturer’s instructions. RNA concentration was determined by measuring the absorbance at 260 nm. Subsequently, 2 μg of RNA was treated with DNase prior to reverse transcription to cDNA using a PrimeScript RT Reagent Kit (Takara; Dalian, China). The cDNA samples were stored at −20 °C.
To measure the mRNA levels of GPx isoforms 1, 3, and 4 (GPx1, GPx3, and GPx4), and SelW, real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed using a 7500 Real Time PCR System (Applied Biosystems; Carlsbad, CA, USA) and SYBR® Premix Ex TaqII Kit (Takara; Dalian, China). The volume of each PCR reaction was 25 μL, which included 2 μL cDNA, 12.5 μL SYBR® Premix Ex TaqII, 1 μL PCR forward primer, 1 μL PCR reverse primer, and 8.5 μL ddH2O. Beta-actin was used as the internal control gene, and the information for all primers is listed in Table 2. All data were normalized to the internal control beta-actin and relative expression levels were calculated using the 2−△△Ct method.
Statistical analysis
Results were analyzed by one-way analysis of variance (ANOVA), followed by a multiple comparison test (least square difference; LSD). Statistical analyses were performed using SPSS statistical software (version 18.0; SPS, Chicago, IL, USA). All data are presented as means ± SEM. P values less than 0.05 were considered to be statistically significant.
Results
Organ weight and Se deposition
During the experimental period, addition of Se to the diet noticeably increased liver organ index in mice treated with 0.4, 0.8, or 0.045 mg Se/kg, compared to mice treated with 0.1 mg Se/kg (P < 0.05; Table 3). A greater increase in kidney organ index was observed in mice dosed with 0.8 mg Se/kg compared with those receiving 0.045 or 0.1 mg Se/kg.
Deposition of Se in the liver and kidneys increased significantly (P < 0.05) with increasing amounts of dietary Se (Table 3). In addition, Se deposition in the kidneys did not differ between mice receiving dietary supplementation with 0.045 or 0.1 mg Se/kg.
Serum biochemistry
Treatment with an oral dose of 0.4 or 0.8 mg Se/kg significantly (P < 0.05) increased levels of BUN and SCr, indicating kidney toxicity, and significantly (P < 0.05) increased serum levels of AST (P < 0.05), demonstrating that liver function was adversely affected (Table 4). Under the same experimental conditions, treatment with 0.045 mg Se/kg did not change the serum levels of any of the biochemical parameters measured, with the exception of BUN, which increased significantly compared to that in mice treated with 0.1 mg Se/kg (P < 0.05). No significant changes in ALT were observed in male mice treated with dietary Se at any of the administered doses.
Antioxidant capacity in the liver
Dietary Se level significantly correlated (P < 0.05) with liver antioxidant parameters, with the highest level of MDA observed in mice fed a diet containing 0.8 mg Se/kg (Table 5). However, activities of GR, CAT, GPx, and SOD in the liver were significantly reduced (P < 0.05) in mice given the Se-excess diet compared to mice given lower amounts of Se. Treatment with dietary Se at 0.4 mg/kg did not change liver activities of antioxidant parameters, with the exception of CAT, which decreased (P < 0.05), when compared with mice treated with 0.1 mg Se/kg. In addition, dietary supplementation with 0.045 mg Se/kg significantly reduced (P < 0.05) liver activities of GPx in the absence of changes in activities of GR or SOD, when compared to mice treated with 0.1 mg Se/kg.
Levels of selenoprotein mRNA in different organs
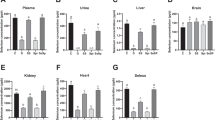
Dietary Se levels exhibited varying effects on the expression of GPx1, GPx3, GPx4, and SelW genes in different organs (Fig. 1). Analysis by qRT-PCR showed that liver GPx1, GPx3 and GPx4 mRNA levels in Se-deficient (0.045 mg/kg) or Se-excess (0.8 mg/kg) mice were decreased (P < 0.0001). Additionally, in mice receiving 0.4 mg Se/kg, liver GPx3 mRNA was decreased and SelW mRNA was increased, while mRNA levels for GPx1 and GPx4 were unchanged, as compared to those in organs from mice fed Se-adequate (0.1 mg/kg) diets. Under the same conditions, there were significant increases in levels of kidney mRNA encoding GPx1, GPx3, GPx4, and SelW in the Se-supernutrition group. No significant differences among dietary groups were observed in the levels of Gpx4 mRNA in mouse testis exposed to 0.045–0.8 mg Se/kg.
Effects of different amounts of dietary selenium on mRNA levels for a subset of selenoproteins in different organs of male BALB/c mice (n = 6) (a, b) Significant difference (P < 0.05) by ANOVA. Values are means ± SEM relative to baseline, group treated with 0.1 mg Se/kg diet. Beta-actin was used as internal control gene for normalization. White bars Se-deficient group (0.045 mg/kg); light gray bars Se-adequate group (0.1 mg/kg); gray bars Se-supernutrition group (0.4 mg/kg); dark gray bars Se-excess group (0.8 mg/kg). Values represent fold change in comparison with the transcription level of the group treated with 0.1 mg Se /kg diet
Discussion
In our current study, to exclude limitations of sex hormone-dependent control of selenoprotein expression [22], male weanling mice were fed diets containing different Se levels. Consistent with several previous studies [23–25], we did not observe a significant effect of dietary Se on growth, even at 0.8 mg/kg body weight, suggesting that Se-dependent effects on growth occur above this level. Additional experiments using higher concentrations of Se should be pursued in the future to examine toxicity.
The dietary requirement for Se is 0.1 mg/kg for mice, according to the National Research Council [26]. In the present study, when mice were fed 0.8 mg Se/kg in the diet, results were in agreement with similar data from rat tissues [25], namely that there were significant increases in liver and kidney Se concentrations, which were 187 and 175 %, respectively, of the levels observed in mice fed a diet of 0.1 mg Se/kg. This is possibly due to selenosis induced by a high dose of inorganic Se. ALT and AST activities in serum are markers of liver damage, and the elevated AST activities suggest that 0.4 mg Se/kg causes modest liver damage.
SCr and BUN levels directly reflect the metabolic status of protein in the body, and are standard indicators of renal damage. In the present study, SCr and BUN levels were both increased when mice were exposed to either 0.4 or 0.8 mg Se/kg, which indicated selenosis induced by Se toxicity in the kidneys. It is noteworthy that the level of BUN, but not that of SCr, was increased in the 0.045 mg Se/kg group as compared to the Se-adequate group. This differential effect on these two parameters suggests that glomerular filtration is normal and creatinine can be readily cleared. Hence, a higher degree of oxidative stress in the kidneys was likely produced in the 0.4 mg Se/kg or 0.8 mg Se/kg diet group, compared to the 0.045 mg Se/kg diet group.
The liver and kidneys are important antioxidant organs in mice. Organ index is a more accurate measure than single organ mass for assessing overall effects. During the experimental period, Se-excess caused an increase in the liver and kidney organ index, which suggested that both liver and kidney damage occurred.
Normal levels of Se present during homeostasis up-regulate endogenous antioxidant defenses by increasing GPx activity [27]. Se acts indirectly as an antioxidant through its incorporation into the enzyme Se-dependent GPx [28]. In the present study, the activity of GPx in the liver increased in animals fed a diet containing 0.4 mg Se/kg, but the activity was decreased when Se levels were increased to 0.8 mg/kg. Dietary Se resulted in dose-dependent increases (P < 0.05) in Se concentrations and GPx activities in the liver. Data from the present study also agree with previous reports [13]. In the group fed with Se at a dose of 0.4 mg/kg, an increase in GPx activity is likely based on a free radical mechanism. This is because at higher concentrations, Se reacts with GSH to form a highly reactive redox species, GSSe–, a selenopersulfide [12], that further reacts with GSH to generate superoxide anion.
However, excess Se intake can result in adverse health effects. Thus, altered GPx levels may expose liver cells to oxidative stress. This study demonstrated that different types of oxidant–antioxidant imbalance may be induced by dietary modulation. It has been reported that Se toxicity was related to the generation of reactive oxygen species [29]. The active site of Se-dependent GPx is a selenocysteine that is incorporated into the enzyme during protein synthesis [30]. Inasmuch as the toxicity of Se is thought to be related to the generation of reactive oxygen species [29], it may be concluded that the process of cell damage induced by excess Se triggers the breakdown of regulatory controls and associated loss of protein synthesis, which is accompanied by the decrease of GPx activity. The MDA levels we observed are consistent with the level of lipid peroxidation (LPO) found in tissues [31]. A highly significant increase in LPO was recorded in the liver of mice fed on both 0.4 and 0.8 mg Se/kg diets, compared to the Se-adequate diet-fed animals. The marked increase in LPO in the 0.8 mg Se/kg diet group can be explained on the basis of decreased levels of GPx, which would lead to peroxide accumulation and oxidative stress.
To investigate the effect of differences in Se status on mRNA expression of antioxidant selenoproteins, we examined the regulation of GPx1, GPx3, GPx4, and SelW. In agreement with the previous study [20], supernutritional Se (0.4 mg/kg) status did not change GPx1 or GPx4 mRNA levels in the liver, but did affect GPx3 and SelW. This suggests that hepatic GPx3 and SelW are more sensitive to Se overexposure than other genes. Indeed, Se overexposure had an adverse effect on the hepatic level of GPx3 mRNA. However, a diet containing 0.4 mg Se/kg increased the expression of all four genes in the kidney. Numerous studies have shown that SelW levels increase in response to dietary Se supplementation. Previous studies have shown that upon Se-depletion, SelW and GPx4 mRNA were decreased in colonic mucosa from rats and in human intestinal Caco-2 cells [32], whereas, SelW mRNA was increased in the tongue from rats and sheep [33, 34]. In rats with a severely Se-deficient diet, Gpx4 mRNA levels were unchanged in the liver and heart compared to animals on normal diets [6]. In our experiment, excess Se (0.8 mg/kg) led to decreased GPx1, GPx3, and GPx4 mRNA levels in the liver, but the mRNA level of GPx4 in the testis were not influenced by dietary Se. In view of the above findings, one can conclude that oxidative toxicity induced by varying Se concentrations modulates the expression of GPx1, GPx3, GPx4, and SelW. This strongly suggests that there is an underlying mechanism involving Se-dependent regulation of selenoprotein mRNA levels, although further studies will be required to elucidate this mechanism.
We suggest there are at least two possible explanations for the effects of Se observed in this study. First, the liver and kidneys are more sensitive to redox imbalance than the testis, consistent with the relatively stable metabolism of Se in the testis. Second, GPx4 is prominent among the antioxidant selenoprotein family of proteins that are affected by Se deficiency or excess, and the mRNA level of this gene exhibits relatively slow changes in response to changes in environmental Se status. For these selenoproteins with Se-regulated mRNA levels, it will also be important to determine if the protein levels of the selenoproteins are also regulated by Se status.
References
Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN (2003) Characterization of mammalian selenoproteomes. Science 300(5624):1439–1443
Li JL, Li HX, Li S, Jiang ZH, Xu SW, Tang ZX (2011) Selenoprotein W gene expression in the gastrointestinal tract of chicken is affected by dietary selenium. Biometals 2 4(2):291–299. doi:10.1007/s10534-010-9395-0
Kaushal N, Bansal MP (2009) Selenium variation induced oxidative stress regulates p53 dependent germ cell apoptosis: plausible involvement of HSP70-2. Eur J Nutr 48(4):221–227. doi:10.1007/s00394-009-0005-2
Gan L, Liu Q, Xu HB, Zhu YS, Yang XL (2002) Effects of selenium overexposure on glutathione peroxidase and thioredoxin reductase gene expressions and activities. Biol Trace Elem Res 89(2):165–175
Fairweather-Tait SJ, Bao Y, Broadley MR, Collings R, Ford D, Hesketh JE, Hurst R (2011) Selenium in human health and disease. Antioxid Redox Signal 14(7):1337–1383
Bermano G, Nicol F, Dyer JA, Sunde RA, Beckett GJ, Arthur JR, Hesketh JE (1995) Tissue-specific regulation of selenoenzyme gene expression during selenium deficiency in rats. Biochem J 311(Pt 2):425–430
Balogh K, Weber M, Erdélyi M, Mézes M (2004) Effect of excess selenium supplementation on the glutathione redox system in broiler chicken. Acta Vet Hung 52(4):403–411
Brigelius-Flohé R, Müller C, Menard J, Florian S, Schmehl K, Wingler K (2001) Functions of GI-GPx: lessons from selenium-dependent expression and intracellular localization. Biofactors 14(1–4):101–106
Sunde RA, Hadley KB (2010) Phospholipid hydroperoxide glutathione peroxidase (Gpx4) is highly regulated in male turkey poults and can be used to determine dietary selenium requirements. Exp Biol Med 235(1):23–31. doi:10.1258/ebm.2009.009262
Barnes KM, Evenson JK, Raines AM, Sunde RA (2009) Transcript analysis of the selenoproteome indicates that dietary selenium requirements of rats based on selenium-regulated selenoprotein mRNA levels are uniformly less than those based on glutathione peroxidase activity. J Nutr 139(2):199–206. doi:10.3945/jn.108.098624
Sunde RA, Raines AM, Barnes KM, Evenson JK (2009) Selenium status highly regulates selenoprotein mRNA levels for only a subset of the selenoproteins in the selenoproteome. Biosci Rep 29(5):329–338. doi:10.1042/BSR20080146
Shen HM, Yang CF, Ding WX, Liu J, Ong CN (2001) Superoxide radical-initiated apoptotic signaling pathway in selenite-treated HEPG2 cells: mitochondria serve as the main target. Free Radic Biol Med 30(1):9–21
Kaushal N, Bansal MP (2009) Diminished reproductive potential of male mice in response to selenium-induced oxidative stress: Involvement of HSP70, HSP70-2, and MSJ-1. J Biochem Mol Toxicol 23(2):125–136. doi:10.1002/jbt.20276
Zoidis E, Pappas AC, Georgiou CA, Komaitis E, Feggeros K (2010) Selenium affects the expression of GPx4 and catalase in the liver of chicken. Comp Biochem Phys B 155(3):294–300. doi:10.1016/j.cbpb.2009.11.017
Zhou JC, Zhao H, Li JG, Xia XJ, Wang KN, Zhang YJ, Liu Y, Zhao Y, Lei XG (2009) Selenoprotein gene expression in thyroid and pituitary of young pigs is not affected by dietary selenium deficiency or excess. J Nutr 139(6):1061–1066. doi:10.3945/jn.109.104901
Shalini S, Bansal MP (2005) Role of selenium in regulation of spermatogenesis: Involvement of activator protein 1. Biofactors 23(3):151–162
Terada A, Yoshida M, Seko Y, Kobayashi T, Yoshida K, Nakada M, Nakada K, Echizen H, Ogata H, Rikihisa T (1999) Active oxygen species generation and cellular damage by additives of parenteral preparations: selenium and sulfhydryl compounds. Nutrition (Burbank, Los Angeles County, Calif) 15(9):651–655
Hoffman DJ, Heinz GH, Krynitsky AJ (1989) Hepatic glutathione metabolism and lipid peroxidation in response to excess dietary selenomethionine and selenite in mallard ducklings. J Toxicol Environ Health 27(2):263–271
Balogh K, Weber M, Erdélyi M, Mézes M (2004) Effect of excess selenium supplementation on the glutathione redox system in broiler chicken. Acta Vet Hung 52(4):403–411
Raines AM, Sunde RA (2011) Selenium toxicity but not deficient or super-nutritional selenium status vastly alters the transcriptome in rodents. BMC Genomics 12:26. doi:10.1186/1471-2164-12-26
Bradford HF, Dodd PR (1977) Convulsions and activation of epileptic foci induced by monosodium glutamate and related compounds. Biochem Pharmacol 26(3):253–254
Stoedter M, Renko K, Hög A, Schomburg L (2010) Selenium controls the sex-specific immune response and selenoprotein expression during the acute-phase response in mice. Biochem J 429(1):43–51. doi:10.1042/BJ20091868
Lei XG, Evenson JK, Thompson KM, Sunde RA (1995) Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J Nutr 125(6):1438–1446
Barnes KM, Evenson JK, Raines AM, Sunde RA (1997) Dietary selenium regulation of glutathione peroxidase mRNA and other selenium-dependent parameters in male rats. J Nutr Biochem 8(2):85–91
Barnes KM, Evenson JK, Raines AM, Sunde RA (2009) Transcript analysis of the selenoproteome indicates that dietary selenium requirements of rats based on selenium-regulated selenoprotein mRNA levels are uniformly less than those based on glutathione peroxidase activity. J Nutr 139(2):199–206. doi:10.3945/jn.108.098624
Subcommittee on Laboratory Animal Nutrition, Committee on Animal Nutrition, Board on Agriculture, National Research Council, National Academy of Sciences (1995) Nutrient requirements of laboratory animals. National Academy Press, Washington
Gan L, Liu Q, Xu HB, Zhu YS, Yang XL (2002) Effects of selenium overexposure on glutathione peroxidase and thioredoxin reductase gene expressions and activities. Biol Trace Elem Res 89(2):165–175
Hoekstra WG (1975) Biochemical function of selenium and its relation to vitamin E. Fed Proc 34:2083–2089
Zhu Z, Kimura M, Itokawa Y, Aoki T, Takahashi JA, Nakatsu S, Oda Y, Kikuchi H (1996) Apoptosis induced by selenium in human glioma cell lines. Biol Trace Elem Res 54(2):123–134
Doni MG, Falanga A, Delaini F, Vicenzi E, Tomasiak M, Donati MB (1984) The effect of vitamin E or selenium on the oxidant-antioxidant balance in rats. Br J Exp Pathol 65(1):75–80
Kang BP, Bansal MP, Mehta U (2000) Hyperlipidemia and type I 5′-monodeiodinase activity: regulation by selenium supplementation in rabbits. Biol Trace Elem Res 77(3):231–239
Pagmantidis V, Bermano G, Villette S, Broom I, Arthur J, Hesketh J (2005) Effects of Se-depletion on glutathione peroxidase and selenoprotein W gene expression in the colon. FEBS Lett 579(3):792–796
Yeh JY, Vendeland SC, Gu Q, Butler JA, Ou BR, Whanger PD (1997) Dietary selenium increases selenoprotein W levels in rat tissues. J Nutr 127(11):2165–2172
Yeh JY, Gu QP, Beilstein MA, Forsberg NE, Whanger PD (1997) Selenium influences tissue levels of selenoprotein W in sheep. J Nutr 127(3):394–402
Author information
Authors and Affiliations
Corresponding author
Additional information
Qin Zhang, Long Chen, Kai Guo, and Zhaoxin Tang contributed equally to this work.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Zhang, Q., Chen, L., Guo, K. et al. Effects of Different Selenium Levels on Gene Expression of a Subset of Selenoproteins and Antioxidative Capacity in Mice. Biol Trace Elem Res 154, 255–261 (2013). https://doi.org/10.1007/s12011-013-9710-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12011-013-9710-z