Abstract
In this study, we studied the biochemical characterization of flavone synthase I from Daucus carota (DcFNS I) and applied it with flavonoid 6-hydroxylase from Scutellaria baicalensis (SbCYP) to convert flavanones to flavones. The recombinant DcFNS I was expressed in the form of the glutathione-S-transferase fusion protein. Rather than taxifolin, naringenin, pinocembrin, and eriodictyol were accepted as substrates. The optimal temperature and pH for reaction in vitro were 35 °C and 7.5, respectively, and 2-oxoglutarate was essential in the assay system. Co2+, Cu2+, Mn2+, Ni2+, and Zn2+ were not substitutes for Fe2+. EDTA and pyruvic acid inhibited the activity, except for Fe3+. Kinetic analysis revealed that the Vmax and kcat values of the recombinant DcFNS I against naringenin were 0.183 nmol mg−1 s−1 and 0.0121 s−1, and 0.175 nmol mg−1 s−1 and 0.0116 s−1 against pinocembrin. However, the recombinant DcFNS I had a higher affinity for naringenin than pinocembrin, with kM values for each of 0.076 mM and 0.174 mM respectively. Thus, it catalyzed naringenin more efficiently than pinocembrin. Subsequently, using an Escherichia coli and Saccharomyces cerevisiae co-culture system, we successfully converted naringenin and pinocembrin to scutellarein and baicalein respectively. In a synthetic complete medium, the titers of scutellarein and baicalein reached 5.63 mg/L and 0.78 mg/L from 200 mg/L precursors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Flavonoids are polyphenolic secondary metabolites produced by most plants [1] and are classified into several groups, such as flavanones, flavones, flavanonols, flavonols, isoflavones, and anthocyanins [2]. They are critical in plant growth and development, such as protection against ultraviolet radiation, resistance to pest invasion, and acting as phytoalexins [3]. Moreover, flavonoids have many benefits for humans and are used in food and medicine [4]. Scutellarein and baicalein are members of flavonoids and are primarily found in the medicine plant Scutellaria baicalensis Georgi, with numerous bioactivities [5]. In particular, baicalein has recently been demonstrated to have an inhibitory effect on SARS-CoV-2 3CL [6]. Considering the application prospect of these compounds, obtaining large amounts of such compounds is a problem to meet the booming demand. Conventional methods, like plant extraction and chemical synthesis, are unsustainable and contaminative. However, microbial synthesis has brought a new advancement for this purpose. Some researches reported that scutellarein and baicalein were successfully produced in engineered microbes, such as Escherichia coli or Pichia pastoris [7, 8].
The entire biosynthetic pathway of flavonoids in plants has been illustrated and is divided into three parts: phenylpropanoid pathway, polyketide synthesis, and post-modification [9]. This pathway consists of a cascade of various enzymes, in which flavone synthase catalyzes the conversion of flavanones into flavones by desaturation reaction. Flavone synthases are divided into two categories, each with a different catalytic mechanism [10]. Flavone synthase I (FNS I) is a dioxygenase and belongs to the 2-oxoglutarate-dependent dioxygenase (2ODD) enzyme family [11]. Flavone synthase II (FNS II) is a monooxygenase and belongs to the membrane-bound cytochrome P450 enzymes family [12]. Many flavone synthases have been found in various plants of different families, such as the Apiaceae, Aytoniaceae, and Oryzoideae [11, 13, 14]. Daucus carota is a member of the Apiaceae family, and acts as a vegetable in our daily diet, which is rich in flavonoids, terpenes, and vitamins. Some genes related to the biosynthetic pathway of flavonoids in this plant have been reported before [15, 16]. However, there are only a few reports that illustrated their detailed biochemical properties, which are useful for producing flavonoids in engineered strains by metabolic engineering or synthetic biology.
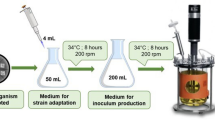
Here, a flavone synthase I from Daucus carota (DcFNS I) was expressed in E.coli. To overcome some problems of plant protein expressed in prokaryotes, such as inclusion body, and to get enough protein within a short time for the study of its biochemical characteristics, DcFNS I was expressed in the form of a glutathione-S-transferase fusion protein because this fusion protein system has many advantages, e.g., time-saving and promoting soluble protein expression [17]. The biochemical characteristics of DcFNS I were described detailedly in this study. After that, we constructed a co-culture system of E. coli and S. cerevisiae to convert naringenin and pinocembrin to scutellarein and baicalein respectively by coupling this enzyme with a flavonoid 6-hydroxylase (F6H) from Scutellaria baicalensis [18] and an NADPH-cytochrome P450 reductase from Arabidopsis thaliana [19] (Fig. 1). Through this consortium, the target products were successfully produced. This was the first time to produce scutellarein and baicalein from their precursors in engineered microbial consortia.
Materials and Methods
Strains, Plasmids, Media, and Chemicals
All strains and plasmids used in this study are listed in Table 1. E. coli DH5α was used for plasmid cloning and propagation. E. coli BL21 (DE3) and S. cerevisiae YPH499 were employed for exogenous protein expression and flavonoid production. Lysogeny broth (LB) and terrific broth (TB) were utilized for E. coli strains cultivation. The former contains 10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl, and the latter contains 11.8 g/L tryptone, 23.6 g/L yeast extract, 9.4 g/L K2PO4, 2.2 g/L KH2PO4, and 0.4% glycerol. SD or SG dropout medium was deployed for cultivating S. cerevisiae strains. The former contains 2% glucose, 0.67% yeast nitrogen without amino acids, and 1.39 g/L Yeast Synthetic Drop-out Media Supplements Y2001 (Sigma), and the latter contains similar ingredients except for 2% d-galactose. The synthetic complete medium for co-incubating E. coli BL21 (DE3) and S. cerevisiae YPH499 was composed of 2% glucose, 0.67% yeast nitrogen without amino acids, and amino acid mixture. The amino acid mixture contained the following: 20 mg/L adenine, 20 mg/L uracil, 20 mg/L l-histidine, 20 mg/L l-arginine, 20 mg/L l-methionine, 30 mg/L l-tyrosine, 100 mg/L l-leucine, 30 mg/L l-isoleucine, 30 mg/L l-lysine, 50 mg/L l-phenylalanine, 150 mg/L l-valine, and 200 mg/L l-threonine. Authentic standard chemicals (99%) were purchased from Plant Origin Biological, Nanjing. Ampicillin (100 mg/L) was added to the medium when required.
Gene Cloning and Plasmid Construction
DcFNS I cDNA (AY817675.1) [16] was synthesized according to the codon preference of E. coli and subcloned into BamHI/EcoRI site of pGEX-4 T. Similarly, SbCYP cDNA (MF363006.1) [18] and ATR2 cDNA (NM_179141.2) [19] were synthesized according to the codon preference of S. cerevisiae and subcloned into EcoRI/NotI site of pESC-Trp and BamHI/XhoI site of pESC-His respectively. pESC-Trp-SbCYP-ATR2(G + G) and pESC-Trp-SbCYP-ATR2(T + P) were constructed by restriction endonucleases. pRS313-His-SbCYP-ATR2(G + G) and pRS313-His-SbCYP-ATR2(T + P) were constructed by in-fusion cloning. Briefly, an amplicon amplified from pESC-Trp-SbCYP-ATR2(G + G) with primers GG-F and GG-R was combined with PCR product amplified from pRS313 with primers pRS-F and pRS-R to construct pRS313-His-SbCYP-ATR2(G + G), and an amplicon amplified from pESC-Trp-SbCYP-ATR2(T + P) with primers sa-F and sa-R was combined with PCR product amplified from pRS313 with primers pRS313-F and pRS313-R to construct pRS313-His-SbCYP-ATR2(T + P). All primers used in this study are listed in Table S1. Restriction endonuclease, DNA polymerase, and T4 DNA ligase were purchased from Takara Biochemicals (Japan). In-fusion cloning kit was purchased from Beyotime Biotechnology Company (Shanghai, China).
Recombinant Protein Expression and Purification
To express the recombinant protein DcFNS I, pGEX-4 T and pGEX-4 T-DcFNSI were transformed into E. coli BL21 (DE3) competent cells by heat shock to form B0 and B1 respectively. The resulting strains were cultivated in a 5-mL LB medium at 37 °C overnight, then 1 ml of seeds was transferred to a 50-mL LB medium. When OD600 reached 0.8, IPTG at a final concentration of 1 mM was added to induce gene expression at 30 °C for 5 h. The cells were collected by centrifugation at 4 °C at 8000 rpm for 10 min and washed twice with deionized water for recombinant protein purification. The recombinant protein DcFNS I was purified using a GST-Sefinose (TM) Kit (BBI Life Science, China), according to the manufacturer’s instructions.
Substrate Specificity
To determine the substrate specificity of the recombinant protein DcFNS I, the frozen glycerol stock of B1 kept at − 80 °C was inoculated into a 5-mL LB medium and growing at 37 °C overnight. The pre-inoculum was inoculated into a 5-mL TB medium until OD600 reached 0.8. IPTG was added at a final concentration of 0.5 mM to induce gene expression at 30 °C for 5 h, 0.5 mM sodium ascorbate, 0.5 mM ferrous ion, 0.5 mM 2-oxoglutarate, and one substrate (200 mg/L) were added. The culture was incubated for another 24 h, and samples were collected for HPLC and HPLC/MS analysis.
Enzyme Assays
The assay system contained 200 μL total volumes, including 100 mM Na2HPO4–KH2PO4 or glycine–NaOH, 10 mM sodium ascorbate, 100 µM ferrous ion, 250 µM 2-oxoglutarate, 50 µM substrate, and 120 µg purified protein DcFNS I. The substances were added in this sequence. After shaking the mixture for 10 s on a vortex mixer, it was incubated in a water bath. The reaction was terminated by adding an equal volume of methanol for HPLC analysis.
The assays were performed at pH 7 for 1 h at temperatures 28, 32, 35, 38, or 42 °C to determine the optimal temperature for the reaction. The thermal stability of the protein was determined by incubating the protein for varying lengths of time at 30, 35, or 40 °C. The assays were performed at 35 °C for 1 h to determine the optimal pH for the reaction using 100 mM of Na2HPO4–KH2PO4 buffer (pH 5.5–8.0) and glycine–NaOH buffer (pH 8.6–10.0). The pH stability of the protein was determined by incubating it at 4 °C for different periods at pH 7, 7.5, or 8. To determine the cofactor requirement, α-oxoglutarate, Fe2+, and sodium ascorbate were evaluated for their necessity in reaction. To investigate the effect of different metal ions and potential inhibitors on the activity, Co2+, Cu2+, Mn2+, Ni2+, Zn2+, Fe3+, EDTA, and pyruvic acid were examined. Naringenin was employed as a substrate for all analyses. To determine kinetic parameters of the recombinant protein DcFNS I, the analyses were performed as follows: substrate concentrations (0.01 ~ 250 μM) of naringenin or pinocembrin, pH (7.5), temperature (35 °C), protein (27 μg), and incubation for 1 or 2 h. KM, Vmax, and kcat values were calculated from nonlinear regression analysis using the software of Origin 9.
Co-culture Fermentation
The co-culture system was divided into two modules. The upstream module was comprised of B1. The downstream module was comprised of Y1, Y2, Y3, or Y4. The plasmids of pESC-Trp-SbCYP-ATR2(G + G), pRS313-His-SbCYP-ATR2(G + G), pESC-Trp-SbCYP-ATR2(T + P), and pRS313-His-SbCYP-ATR2(T + P) were transformed into S. cerevisiae YPH499 competent cells by electroporation to form strains Y1, Y2, Y3, and Y4 respectively. For screening the optimal downstream module, the four strains were pre-cultured in SD-Trp or SD-His dropout medium for 24 h, then inoculated into a 25 mL SD-His or SG-His dropout medium supplemented with 100 mg/L apigenin or chrysin, with an initial OD600 of 0.05, and incubated at 30 °C for 5 days. The samples were collected at the end of fermentation for HPLC analysis.
Before co-culture fermentation, two seeds were prepared as follows. B1 was cultured overnight in a 5-mL LB medium at 37 °C, then pre-inoculum was inoculated into a 40-mL LB medium until OD600 reached 0.8. IPTG was added at a final concentration of 1 mM. The culture was incubated at 30 °C for 6 h before harvest. Y4 was cultured in a 5-mL SD-His medium at 30 °C for 24 h, then 1% inoculum was inoculated into a 40-mL SD-His medium. Before harvest, the culture was incubated at 30 °C for 24 h. Two prepared seeds were transferred to a synthetic complete medium supplemented with naringenin or pinocembrin for co-culture fermentation. The factors of inoculation ratios (9:1, 4:1, 1:1, 1:4, and 1:9) with a total OD600 of 4.0, temperature (25 °C, 30 °C, and 35 °C), and substrate concentration (50, 100, 200, 400, and 800 mg/L) were systematically evaluated to promote the titer of the target products.
Analytical Methods
BioPhotometer plus (Eppendorf, Germany) was used to measure optical density at 600 nm. For analyzing flavonoids in the fermentation broth, 500 µL culture was diluted with an equal volume of methanol. After vigorous mixing and ultrasonication for 10 min, the lysates were spun down at 12,000 rpm for 10 min. The supernatant was filtered through a 0.45-µm membrane before measurement. The samples (20 μL) were analyzed by Agilent 1260 Infinity system equipped with a photodiode-array detector. The products were detected at 270 nm, 290 nm, and 340 nm using a reverse-phase C18 column (5 µm, 250 × 4.6 nm) operating at 30 °C. The solvents were (A) methanol and (B) water containing 0.1% formic acid at a flow rate of 0.8 mL/min. The separation program used was as follows: 0–20 min, 30–80% A; 20–25 min, 80–100% A; 25–32 min, 100–30% A.
Results and Discussion
Expression and Purification of the Recombinant Protein DcFNS I
Here, a flavone synthase I from Daucus carota was expressed in E. coli BL21 (DE3) as a form of the glutathione-S-transferase fusion protein. The recombinant DcFNS I in B1’s lysate was purified by using GST columns and showed near homogeneity on an SDS-PAGE gel (Fig. 2). The fusion protein had a molecular weight of 66.2 kDa, corresponding to the sum of GST (26 kDa) and DcFNS I (40.2 kDa). This molecular weight was comparable to other FNS I, such as Plagiochasma appendiculatum (41.02 kDa) [14] and Conium maculatum (40.96 kDa) [16]. B0 carrying empty vector pGEX-4 T was used as a control.
Expression and purification of the recombinant protein DcFNS I. Lane 1, standard protein markers. Lane 2, the supernatant fraction of strain B1 after cell disruption and centrifugation. Lane 3, the supernatant fraction of strain B0 after cell disruption and centrifugation. Lane 4, the purified protein of DcFNS I. Lane 5, the purified protein of strain B0 (empty control)
Substrate Specificity of DcFNS I
The substrate specificity of DcFNS I was determined. When flavanone compounds, naringenin, pinocembrin, and eriodictyol, were fed as substrates, three products were detected by HPLC analysis, identical to retention times of authentic standards, apigenin, chrysin, and luteolin respectively. No products but only substrates were detected in the fermentation broth of the empty control (Fig. S1). We further performed LC–MS analysis to investigate the molecular mass of these products. The products had a mass-to-charge ratio (m/z) of 271.06, 255.06, and 287.05 [M + H]+ respectively, which were identical to those generated from authentic standards (Fig. S2). These results confirmed that the recombinant protein DcFNS I catalyzed the dehydrogenation of 2, 3 carbon atoms in the C ring of flavanones to corresponding products. When taxifolin, a flavanonol compound, was used as the substrate, no product was detected by HPLC analysis (data not shown). Some research results also drew a parallel conclusion [11]. However, PaFNS I showed some flavonol synthase activity by converting dihydrokaempferol to kaempferol in vitro [14]. This is of interest because a novel function of FNS I was reported.
Effect of Temperature and pH on the Activity and Stability of DcFNS I
The optimal temperature for the reaction was 35 °C in vitro, while the activity was maintained at least 85% in the range of 28 to 42 °C (Fig. 3a). The results demonstrated that temperature affects the activity of DcFNS I slightly in this range. The thermal stability was examined at 30, 35, and 40 °C respectively. The results revealed that its residual activity decreased significantly after 4 h of incubation at 40 °C, while more than 75% was kept after being incubated at 30 or 35 °C for the same duration (Fig. 3c). The optimal pH for the reaction was 7.5, with at least 80% of the activity remaining in the range of 7 to 9. However, the activity decreased to approximately 15% at pH 10 (Fig. 3b). The pH stability was examined at pH 7, 7.5, and 8 respectively. The results have shown that its residual activity remained above 80% after being incubated at 4 °C at different pH for 12 h (Fig. 3d).
Cofactor Requirement of DcFNS I
The cofactor requirement of DcFNS I was investigated. As depicted in Table 2, 2-oxoglutarate was essential for the activity, while removing Fe2+ or sodium ascorbate resulted in the activity degeneration. DcFNS I lost complete activity when three cofactors were removed from the assay system. There was a paradox that the activity of DcFNS I was detected in the absence of Fe2+ in vitro. In fact, molecular oxygen, 2-oxoglutarate, and Fe2+ are necessities for this enzyme, except for ascorbate. The former activates with a flavone synthase I to oxidative dehydrogenation of the substrates to format the corresponding products, and the latter acts as a reductant only [20]. Therefore, the probable reason may be due to the contamination of Fe2+ during the purification process of DcFNS I.
Effect of Metal Ions and Potential Inhibitors on the Activity
Several divalent metal ions were tested for investigating their possibility of substitution of Fe2+. As shown in Table 3, unfortunately, none of them was competent. Among these ions, Co2+ even resulted in the activity being completely lost. EDTA (2.5 mM), pyruvic acid (20 mM), and Fe3+ (100 μM) were also tested as potential inhibitors. The results revealed that EDTA and pyruvic acid were both strong inhibitors. EDTA is a metal ion chelating agent which possibly chelates Fe2+ in the assay system to hinder reaction, and pyruvic acid is an analog of 2-oxoglutarate which may compete with 2-oxoglutarate to occupy the pocket of the catalytic active center of FNS I, while Fe3+ had a minor positive effect on the activity.
Kinetic Analysis
Table 4 presented the kinetic parameters of DcFNS I. The apparent constant KM and Vmax for naringenin were calculated to be 76 μM and 0.183 nmol mg−1 s−1, and 174 μM and 0.175 nmol mg−1 s−1 for pinocembrin respectively. The kcat values for naringenin and pinocembrin were calculated as 0.0121 s−1 and 0.0116 s−1 respectively. The results revealed that the recombinant protein had a higher catalytic efficiency (kcat/KM) for naringenin than pinocembrin.
Screening for the Best Downstream Module
We constructed four different strains to select the best downstream module. In comparison to Y1, the genes of SbCYP and ATR2 in Y2 were controlled under the same promoters but had lower copy numbers. The same situation was for Y3 and Y4. Y4 achieved the highest conversion efficiency for apigenin and chrysin, with titers of 9.6 mg/L and 7.2 mg/L for scutellarein and baicalein respectively. Y1 showed the lowest conversion efficiency, with scutellarein and baicalein titers of 2.3 mg/L and 3.9 mg/L respectively. The conversion efficiency of Y2 and Y3 was in between that of Y1 and Y4 (Fig. S3). Given the above results, we chose B1 and Y4 to construct the co-culture system.
Co-culture for the Production of Scutellarein and Baicalein
After two seeds were prepared, we evaluated the possibility of the co-culture system to produce flavones from flavanones. B1 and Y4 were mixed at a 1:1 ratio and incubated at 30 °C. After 5 days, we detected four products under that condition (Fig. S4). When naringenin was used as the precursor, the intermediate product apigenin and the final product scutellarein titers were 39.7 and 3.42 mg/L respectively. When pinocembrin was added as the precursor, the titers of the intermediate product chrysin and the final product baicalein were 16.97 and 0.78 mg/L respectively. Given the trace amount of baicalein produced from 200 mg/L pinocembrin, we chose naringenin to further investigate the effect of factors on the titer of target products.
The final fermentation results of this E. coli-S. cerevisiae co-culture systems are influenced by fermentation conditions. As expected, the titers of apigenin and scutellarein were significantly influenced by the inoculation ratio of B1 and Y1. The apigenin titer increased along with the increasing relative ratio of B1, achieving 64.9 mg/L in the ratio of 1:9 (S. cerevisiae: E. coli) from 200 mg/L naringenin. However, the higher titer of the intermediate product did not mean a higher titer of the final product. The highest titer of scutellarein reached 5.3 mg/L in an inoculation ratio of 4:1, which was 2.6-fold higher than that in an inoculation ratio of 1:9. Thus, the 4:1 ratio was optimal in this consortium (Fig. 4a). Fermentation temperature was another important factor that influenced the titer of products because S. cerevisiae and E. coli had different optimal temperatures for growth and metabolism. We measured the product’s titer at three different culture temperatures. The highest titer of scutellarein reached 5.63 mg/L at 30 °C, which was slightly higher than that at 25 °C. But, when the temperature was up to 35 °C, the scutellarein titer decreased to 4.43 mg/L. Simultaneously, the apigenin titer was also declining. Therefore, 30 °C was the optimal culture temperature for this consortium (Fig. 4b). We further evaluated the effect of precursor concentration on the titer of products by varying naringenin concentrations from 50 to 800 mg/L. When the concentration of naringenin was 100 mg/L, the titers of apigenin and scutellarein were 13.53 and 5.61 mg/L, respectively, which was nearly twofold higher than that of adding 50 mg/L of naringenin. Nevertheless, the titer of scutellarein no longer increased when the concentration of naringenin was up to 200 mg/L (Fig. 4c). There was a strange phenomenon when the concentration of naringenin reached 400 mg/L and more, we did not detect any products but substrate in these conditions. From the above results, the scutellarin titer enhanced from 3.42 mg/L to the maximum of 5.63 mg/L after changing the fermentation conditions. However, the scutellarin titer was still low. Except for the fermentation conditions, internal factors probably also affect the titer of the target products produced by this consortium. We speculated that the downstream module was more likely to be a rate-limiting step partly due to the instability of the free plasmid that was introduced into S. cerevisiase. Consequently, the genes SbCYP and ATR2 were integrated into the chromosome of S. cerevisiase to construct a downstream module that was more stable in genetics. However, that was not useful (data not shown). Many other internal factors could result in the low titer, e.g., the incompatibility of the P450 and CPR [21], the insufficient pool of reduced coenzyme II (NADPH) in engineered S. cerevisiae [22], the low mass transfer efficiency between strains, and the low catalytic efficiency of FNS I and F6H. These need to be examined and the process is time-costing.
In conclusion, we primarily systematically characterized a flavone synthase I (DcFNS I) from the plant Daucus carota, providing a better understanding of this type of enzyme. Next, an E. coli-S. cerevisiae co-culture system was constructed to produce scutellarein and baicalein from naringenin and pinocembrin respectively, in which the engineered E. coli carried DcFNS I to form the upstream module, and the engineered S. cerevisiae carried SbCYP and ATR2 to form the downstream module. Scutellarein and baicalein were successfully produced by this consortium, but the titers of the target products were unsatisfactory. Much work needs to be done to improve the titers. Nonetheless, these findings presented the feasibility of producing scutellarein and baicalein from their precursors by coculturing E. coli and S. cerevisiae.
Data Availability
Data supporting the productivity of this investigation are available from the corresponding author upon request.
Abbreviations
- FNS I:
-
Flavone synthase I
- FNS II:
-
Flavone synthase II
- 2ODDs:
-
2-Oxoglutarate-dependent dioxygenases
- DcFNS I:
-
Flavone synthase I from Daucuscarota
- SbCYP:
-
P450 hydroxylase from Scutellariabaicalensis
- ATR2:
-
NADPH-cytochrome P450 reductases from Arabidopsis thaliana
References
Kumar, M., Dahuja, A., Tiwari, S., Punia, S., Tak, Y., Amarowica, R., Bhoite, A. G., Singh, S., Joshi, S., Panesar, P. S., Saini, R. P., Pihlanto, A., Tomar, M., Sharifi-Rad, J., & Kaur, C. (2021). Recent trends in extraction of plant bioactives using green technologies: A review. Food Chemistry, 353, 129431.
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology, 126, 485–493.
Mierziak, J., Kostyn, K., & Kulma, A. (2014). Flavonoids as important molecules of plant interactions with the environment. Molecules, 19, 16240–16265.
Elizabeth, D. L. T. J., Gassara, F., Kouassi, A. P., Bar, S. K., & Belkacemi, K. (2017). Spice use in food: Properties and benefits. Critical Reviews in Food Science and Nutrition, 57, 1078–1088.
Zhao, Q., Chen, X. Y., & Martin, C. (2016). Scutellaria baicalensis, the golden herb from the garden of Chinese medicinal plants. Science Bulletin, 61(18), 1391–1398.
Jo, S., Kim, S., & Kim, M. S. (2020). Inhibition of SARS-CoV 3CL protease by flavonoids. Journal of Enzyme Inhibition and Medicinal Chemistry, 35, 145–151.
Li, J. H., Tian, C. F., Xia, Y. H., Mytanda, I., Wang, K. B., & Wang, Y. (2019). Production of plant-specific flavones baicalein and scutellarein in an engineered E-coli from available phenylalanine and tyrosine. Metabolic Engineering, 52, 124–133.
Qian, Z. L., Yu, J. H., Chen, X. J., Kang, Y. J., Ren, Y. N., Liu, Q., Lu, J., & Zhang, Q. (2022). De novo production of plant 4’-deoxyflavones baicalein and oroxylin A from ethanol in crabtree-negative yeast. ACS Synthetic Biology, 11(4), 1600–1612.
Nabavi, S. M., Samec, D., Tomczyk, M., Milella, L., Russo, D., Habtemariam, S., Suntar, I., Rastrelli, L., Daglia, M., Xiao, J. B., Giampieri, F., Battino, M., Sobarzo-Sanchez, E., Nabavi, S. F., Yousefi, B., Jeandet, P., Xun, S. W., & Shirooie, S. (2020). Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnology Advances, 38, 107316.
Martens, S., & Mithofer, A. (2006). Flavones and flavone synthases. Phytochemistry, 67, 521–521.
Martens, S., Forkmann, G., Matern, U., & Lukacin, R. (2001). Cloning of parsley flavone synthase I. Phytochemistry, 58, 43–46.
Akashi, T., Fukuchi-Mizutani, M., Aoki, T., Ueyama, Y., Yonekura-Sakakibara, K., Tanaka, Y., Kusumi, T., & Ayabe, S. (1999). Molecular cloning and biochemical characterization of a novel cytochrome P450, flavone synthase II, that catalyzes direct conversion of flavanones to flavones. Plant and Cell Physiology, 40, 1182–1186.
Lee, Y. J., Kim, J. H., Kim, B. G., Lim, Y., & Ahn, J. H. (2008). Characterization of flavone synthase I from rice. BMB Reports, 41, 68–71.
Han, X., Wu, Y. F., Gao, S., Yu, H. N., Xu, R. X., Lou, H. X., & Cheng, A. X. (2014). Functional characterization of a Plagiochasma appendiculatum flavone synthase I showing flavanone 2-hydroxylase activity. FEBS Letters, 588, 2307–2314.
Hirner, A. A., Veit, S., & Seitz, H. U. (2014). Regulation of anthocyanin biosynthesis in UV-A-irradiated cell cultures of carrot and in organs of intact carrot plants. Plant Science, 161(2), 315–322.
Gebhardt, Y., Witte, S., Forkmann, G., Lukacin, R., Matern, U., & Martens, S. (2005). Molecular evolution of flavonoid dioxygenases in the family Apiaceae. Phytochemistry, 66, 1273–1284.
Smith, D. B., & Johnson, K. S. (1988). Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene, 67(1), 31–34.
Zhao, Q., Cui, M. Y., Levsh, O., Yang, D. F., Liu, J., Li, J., Hill, L., Yang, L., Hu, Y. H., & Weng, J. K. (2018). Two CYP82D Enzymes function as flavone hydroxylases in the biosynthesis of root-specific 4 ’-deoxyflavones in Scutellaria baicalensis. Molecular Plant, 11, 135–148.
Urban, P., Mignotte, C., Kazmaier, M., Delorme, F., & Pompon, D. (1997). Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. Journal of Biological Chemistry, 272, 19176–19186.
Britsch, L. (1990). Purification and characterization of flavone synthase I, a 2-oxoglutarate-dependent desaturase. Archives of Biochemistry and Biophysics, 282, 152–160.
Milne, N., Thomsen, P., Mølgaard Knudsen, N., Rubaszka, P., Kristensen, M., & Borodina, I. (2020). Metabolic engineering of Saccharomyces cerevisiae for the de novo production of psilocybin and related tryptamine derivatives. Metabolic Engineering, 60, 25–36.
Paramasivan, K., & Mutturi, S. (2017). Regeneration of NADPH coupled with HMG-CoA Reductase activity increases squalene synthesis in Saccharomyces cerevisiae. Journal of Agriculture and Food Chemistry, 65(37), 8162–8170.
Funding
This work was supported by the National Key R & D Program of China (2016YFD0600805) and the Forestry Achievements of Science and Technology to Promote Projects ([2017]10).
Author information
Authors and Affiliations
Contributions
X. Z. designed and executed the experiments and drafted the manuscripts. Z. Q., X. F., and H. Y. interpreted the analytical data. J. P. and L. Z. helped to revise the manuscript. All authors have approved the manuscript in the current form.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent for Participate
All authors have their consent to publish their work.
Consent for Publication
All authors have their consent to publish their work.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Qi, Z., Fan, X. et al. Biochemical Characterization of a Flavone Synthase I from Daucus carota and its Application for Bioconversion of Flavanones to Flavones. Appl Biochem Biotechnol 195, 933–946 (2023). https://doi.org/10.1007/s12010-022-04176-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-022-04176-0