Abstract
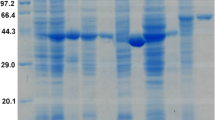
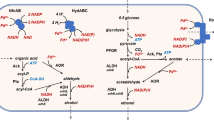
Two iron-containing alcohol dehydrogenases (ADHs) are encoded in the genome of the hyperthermophilic euryarchaeon Thermococcus barophilus Ch5 (Tba ADH641 and Tba ADH547). In our previous publication, we reported biochemical characteristics and catalytic mechanism of Tba ADH547. Herein, we present evidence that Tba ADH641 possesses two activities for ethanol oxidization and acetaldehyde reduction at high temperature, capable of using NAD(H) and NADP(H) as coenzyme. Biochemical data show that Tba ADH641 possesses optimal reaction temperature, thermostability, divalent ion requirement, and substrate specificity distinct from Tba ADH547 and other iron-containing ADH homologues. However, Tba ADH641 and Tba ADH547 display same optimal reaction pH. Kinetic analyses demonstrate that Tba ADH641 displays higher catalytic efficiency for acetaldehyde reduction than that for ethanol oxidation, which is consistent with Tba ADH547. Mutational data demonstrate that residues D115, K118, E159, D190, and E215 in Tba ADH641, which has not been described to date, are necessary for enzyme activity, thus augmenting our understanding on catalytic mechanism of iron-containing ADH. Overall, our work demonstrates that Tba ADH641 is an iron-containing ADH with novel features, which is distinct from Tba ADH547, thus providing a potential biocatalyst for biotransformation reaction.








Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article.
References
Radianingtyas, H., & Wright, P. C. (2003). Alcohol dehydrogenases from thermophilic and hyperthermophilic archaea and bacteria. FEMS Microbiology Reviews, 27(5), 593–616.
Neale, D., Scopes, R. K., Delly, J. M., & Wettenhall, R. E. H. (1986). The two alcohol dehydrogenases of Zymomonas mobilis: Purification by differential dye ligand chromatography, molecular characterization and physiological roles. European Journal of Biochemistry, 154(1), 119–124.
Youngleson, J. S., Jones, W. A., Jones, D. T., & Woods, D. R. (1989). Molecular analysis and nucleotide sequence of the adh1 gene encoding an NADPH-dependent butanol dehydrogenase in the gram-positive anaerobe Clostridium acetobutylicum. Gene, 78(2), 355–364.
De Vries, G. E., Arfman, N., Terpstra, P., & Dijkhuizen, L. (1992). Cloning, expression, and sequence analysis of the Bacillus methanolicus C1 methanol dehydrogenase gene. Journal of Bacteriology, 174(16), 5346–5353.
Conway, T., & Ingram, L. O. (1989). Similarity of Escherichia coli propanediol oxidoreductase (fucO product) and an unusual alcohol dehydrogenase from Zymomonas mobilis and Saccharomyces cerevisiae. Journal of Bacteriology, 171(7), 3754–3759.
Walter, K. A., Bennett, G. N., & Papoutsakis, E. T. (1992). Molecular characterization of two Clostridium acetobutylicum ATCC 824 butanol dehydrogenase isozyme genes. Journal of Bacteriology, 174(22), 7149–7158.
Gaona-López, C., Julián-Sánchez, A., & Riveros-Rosas, H. (2016). Diversity and evolutionary analysis of iron-containing (type-III) alcohol dehydrogenases in eukaryotes. PLoS ONE, 11(11), e0166851.
Williamson, V. M., & Paquin, C. E. (1987). Homology of Saccharomyces cerevisiae ADH4 to an iron-activated alcohol dehydrogenase from Zymomonas mobilis. Molecular and General Genetics, 209(2), 374–381.
Deng, Y., Wang, Z., Gu, S., Ji, C., Ying, K., Xie, Y., et al. (2002). Cloning and characterization of a novel human alcohol dehydrogenase gene (ADHFe1). DNA sequence: The journal of DNA sequencing and mapping, 13(5), 301–306.
Ma, K., Robb, F. T., & Adams, M. W. (1994). Purification and characterization of NADP-specific alcohol dehydrogenase and glutamate dehydrogenase from the hyperthermophilic archaeon Thermococcus litoralis. Applied Environmental Microbiololgy, 60(2), 562–568.
Zhang, L., Jiang, D., Li, Y., Wu, L., Liu, Q., Dong, K., et al. (2021). Characterization of a novel type III alcohol dehydrogenase from Thermococcus barophilus Ch5. International Journal of Biological Macromolecules, 171, 491–501.
Antoine, E., Rolland, J. L., Raffin, J. P., & Dietrich, J. (1999). Cloning and over-expression in Escherichia coli of the gene encoding NADPH group III alcohol dehydrogenase from Thermococcus hydrothermalis. Characterization and comparison of the native and the recombinant enzymes. European Journal of Biochemistry, 264(3), 880–889.
Ma, K., Loessner, H. J., Heider, J., Johnson, M. K., & Adams, M. W. (1995). Effects of elemental sulfur on the metabolism of the deep-sea hyperthermophilic archaeon Thermococcus strain ES-1: Characterization of a sulfur-regulated, non-heme iron alcohol dehydrogenase. Journal of Bacteriology, 177(16), 4748–4756.
Ying, X., Grunden, A. M., Nie, L., Adams, M. W., & Ma, K. (2009). Molecular characterization of the recombinant iron-containing alcohol dehydrogenase from the hyperthermophilic Archaeon Thermococcus strain ES1. Extremophiles, 13(2), 299–311.
Li, D., & Stevenson, K. J. (1997). Purification and sequence analysis of a novel NADP(H)-dependent type III alcohol dehydrogenase from Thermococcus strain AN1. Journal of Bacteriology, 179(13), 4433–4437.
Larson, S. B., Jones, J. A., & McPherson, A. (2019). The structure of an iron-containing alcohol dehydrogenase from a hyperthermophilic archaeon in two chemical states. Acta Crystallographica Section F Structural Biology Communications, 75(Pt 4), 217–226.
Ma, K., & Adams, M. W. W. (1999). An unusual oxygen-sensitive, iron and zinc-containing alcohol dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. Journal of Bacteriology, 181(4), 1163–1170.
Wang, Q., Sha, C., Wang, H., Ma, K., Wiegle, J., Abomohra, A. E., et al. (2021). A novel bifunctional aldehyde/alcohol dehydrogenase catalyzing reduction of acetyl-CoA to ethanol at temperatures up to 95°C. Scientific Reports, 11(1), 1050.
Montella, C., Bellsolell, L., Pérez-Luque, R., Badía, J., Baldoma, L., Coll, M., et al. (2005). Crystal structure of an iron-dependent group III dehydrogenase that interconverts L-lactaldehyde and L-1,2-propanediol in Escherichia coli. Journal of Bacteriology, 187(14), 4957–4966.
Moon, J., Lee, H., Park, S., Song, J., Park, M., Park, H., et al. (2011). Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor. Journal of Molecular Biology, 407(3), 413–424.
Hitschler, L., Nissen, L. S., Kuntz, M., & Basen, M. (2021). Alcohol dehydrogenases AdhE and AdhB with broad substrate ranges are important enzymes for organic acid reduction in Thermoanaerobacter sp. strain X514. Biotechnology for Biofuels, 14(1), 187.
Marteinsson, V. T., Birrien, J. L., Reysenbach, A. L., Vernet, M., Marie, D., Gambacorta, A., et al. (1999). Thermococcus barophilus sp. nov., a new barophilic and hyperthermophilic archaeon isolated under high hydrostatic pressure from a deep-sea hydrothermal vent. International Journal of Systematic Bacteriology, 49(Pt 2), 351–359.
Oger, P., Sokolova, T. G., Kozhevnikova, D. A., Taranov, E. A., et al. (2016). Complete genome sequence of the hyperthermophilic and piezophilic archaeon Thermococcus barophilus Ch5, capable of growth at the expense of hydrogenogenesis from carbon monoxide and formate. Genome Announcements, 4(1), e01534-e1615.
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., de Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Research, 40(Web Server issue), W597-603.
Ying, X., Wang, Y., Badiei, H., Karanassios, V., & Ma, K. (2007). Purification and characterization of an iron-containing alcohol dehydrogenase in extremely thermophilic bacterium Thermotoga hypogea. Archives of Microbiology, 187(6), 499–510.
Elleuche, S., Fodor, K., Klippel, B., voner, H. A., Wilmanns, M., Antranikian G. (2013). Structural and biochemical characterisation of a NAD(+)-dependent alcohol dehydrogenase from Oenococcus oeni as a new model molecule for industrial biotechnology applications. Applied Microbiology and Biotechnology, 97(20): 8963–8975.
Elleuche, S., Fodor, K., von der, H. A., Klippel, B., Wilmanns, M., Antranikian, G. (2014). Group III alcohol dehydrogenase from Pectobacterium atrosepticum: insights into enzymatic activity and organization of the metal ion-containing region. Applied Microbiology and Biotechnology, 98(9): 4041–4051.
Extance, J., Crennell, S. J., Eley, K., Cripps, R., Hough, D. W., & Danson, M. J. (2013). Structure of a bifunctional alcohol dehydrogenase involved in bioethanol generation in Geobacillus thermoglucosidasius. Acta Crystallographica Section D, Structural Biology, 69(Pt 10), 2104–2115.
Sulzenbacher, G., Alvarez, K., Van Den Heuvel, R. H., Versluis, C., Spinelli, S., Campanacci, V., et al. (2004). Crystal structure of E. coli alcohol dehydrogenase YqhD: evidence of a covalently modified NADP coenzyme. Journal of Molecular Biology, 342(2), 489–502.
Acknowledgements
We thank Prof. Philippe Oger at Université de Lyon, INSA de Lyon, for providing the genomic DNA of T. barophilus Ch5.
Funding
This work was supported by the Natural Science Foundation of Jiangsu Province (No. BK20191219), High Level Talent Support Program of Yangzhou University and the Academic Leader of Middle and Young People of Yangzhou University Grant to L.Z, and the Postgraduate Practice Innovation Grant of Jiangsu Province, China (No. SJCX21_1572) to L.W.
Author information
Authors and Affiliations
Contributions
LZ designed experiments; LW performed experiments; LZ and LW analyzed data; and LZ wrote and revised the paper.
Corresponding author
Ethics declarations
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, L., Zhang, L. Biochemical and Functional Characterization of an Iron-Containing Alcohol Dehydrogenase from Thermococcus barophilus Ch5. Appl Biochem Biotechnol 194, 5537–5555 (2022). https://doi.org/10.1007/s12010-022-04052-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-022-04052-x