Abstract
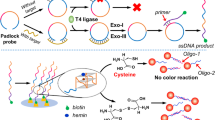
MicroRNAs (miRNAs) play crucial roles in regulating various biological processes and are considered promising biomarkers for clinical diagnosis and therapy of acute pancreatitis. Herein, we present a duplex-specific nuclease (DSN enzyme) and DNAzyme-assisted fluorescent miRNA detections assay that can provide improved detection specificity due to a design of dual-target recognition and a comparable sensitivity. The dual-target recognitions are composed of (i) miRNA unfold hairpin structure toehold to form DNA-RNA duplex, among which the DNA section will be digested by DSN enzyme, releasing miRNA to participant in a next recycle. (ii) After DNAzyme-based nicking site formation in loop section of molecular beacon (MB), miRNA can bind with the loop section of MB and gradually unfold MB probe, generating fluorescence signals. With this general principle, distinct discrimination capability towards even one base pair mismatch of homogenous miRNA is obtained, showing a promising prospect in clinical diagnosis and therapy of acute pancreatitis.
Graphical Abstract





Similar content being viewed by others
Data Availability
All data generated and analyzed during this study are included in this article.
References
Sinonquel, P., Laleman, W., & Wilmer, A. (2021). Advances in acute pancreatitis. Current Opinion in Critical Care, 27, 193–200.
Gariepy, C. E., Ooi, C. Y., Maqbool, A., & Ellery, K. M. (2021). Demographics and risk factors for pediatric recurrent acute pancreatitis. Current Opinion in Gastroenterology, 37, 491–497.
Baeza-Zapata, A. A., Garcia-Compean, D., Jaquez-Quintana, J. O., & Collaborators. (2021). Acute Pancreatitis in Elderly Patients. Gastroenterology, 161, 1736–1740.
Shen, Q., & Reedijk, M. (2021). Notch Signaling and the Breast Cancer Microenvironment. Advances in Experimental Medicine and Biology, 1287, 183–200.
Sabit, H., Cevik, E., Tombuloglu, H., Abdel-Ghany, S., Tombuloglu, G., & Esteller, M. (2021). Triple negative breast cancer in the era of miRNA. Critical Reviews in Oncology/Hematology, 157, 103196.
Backes, C., Meese, E., & Keller, A. (2016). Specific miRNA disease biomarkers in blood, serum and plasma: Challenges and prospects. Molecular Diagnosis and Therapy, 20, 509–518.
Tiwari, A., Mukherjee, B., & Dixit, M. (2018). MicroRNA key to angiogenesis regulation: MiRNA biology and therapy. Current Cancer Drug Targets, 18, 266–277.
Lakshmi, S., Hughes, T. A., & Priya, S. (2021). Exosomes and exosomal RNAs in breast cancer: A status update. European Journal of Cancer, 144, 252–268.
Sukumar, J., Gast, K., Quiroga, D., Lustberg, M., & Williams, N. (2021). Triple-negative breast cancer: promising prognostic biomarkers currently in development. Expert Review of Anticancer Therapy, 21, 135–148.
Zhang, M., Bai, X., Zeng, X., Liu, J., Liu, F., & Zhang, Z. (2021). circRNA-miRNA-mRNA in breast cancer. Clinica Chimica Acta, 523, 120–130.
Chen, C., Ridzon, D. A., Broomer, A. J., Zhou, Z., Lee, D. H., Nguyen, J. T., et al. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Research, 33, e179.
Forero, D. A., Gonzalez-Giraldo, Y., Castro-Vega, L. J., & Barreto, G. E. (2019). qPCR-based methods for expression analysis of miRNAs. Biotechniques, 67, 192–199.
Takei, F., Akiyama, M., Murata, A., Sugai, A., Nakatani, K., & Yamashita, I. (2020). RT-Hpro-PCR: A microRNA detection system using a primer with a DNA tag. Chembiochem, 21, 477–480.
Bejerano, T., Etzion, S., Elyagon, S., Etzion, Y., & Cohen, S. (2018). Nanoparticle delivery of miRNA-21 mimic to cardiac macrophages improves myocardial remodeling after myocardial infarction. Nano Letters, 18, 5885–5891.
Li, F., Li, G., Cao, S., Liu, B., Ren, X., Kang, N., & Qiu, F. (2021). Target-triggered entropy-driven amplification system-templated silver nanoclusters for multiplexed microRNA analysis. Biosensors and Bioelectronics, 172, 112757.
Song, W., Zhu, K., Cao, Z., Lau, C., & Lu, J. (2012). Hybridization chain reaction-based aptameric system for the highly selective and sensitive detection of protein. Analyst, 137, 1396–1401.
Song, Y., Zhang, C., Zhang, J., Jiao, Z., Dong, N., Wang, G., et al. (2019). Localized injection of miRNA-21-enriched extracellular vesicles effectively restores cardiac function after myocardial infarction. Theranostics, 9, 2346–2360.
Uso, M., Jantus-Lewintre, E., Sirera, R., Bremnes, R. M., & Camps, C. (2014). miRNA detection methods and clinical implications in lung cancer. Future Oncology, 10, 2279–2292.
Wu, H., Zhou, W. J., Liu, L., Fan, Z., Tang, H., Yu, R. Q., & Jiang, J. H. (2020). In vivo mRNA imaging based on tripartite DNA probe mediated catalyzed hairpin assembly. Chemical Communications, 56, 8782–8785.
Xu, S., & Lou, Z. (2021). Ultrasensitive detection of nasopharyngeal carcinoma-related miRNA through garland rolling circle amplification integrated catalytic hairpin Assembly. ACS Omega, 6, 6460–6465.
Zhang, G. Z. L., Tong, J., Zhao, X., Ren, J. (2020). CRISPR-Cas12a enhanced rolling circle amplification method for ultrasensitive miRNA detection. Microchemical Journal, 158, 105239.
Zhang, W., Xu, H., Zhao, X., Tang, X., Yang, S., Yu, L., et al. (2020). 3D DNA nanonet structure coupled with target-catalyzed hairpin assembly for dual-signal synergistically amplified electrochemical sensing of circulating microRNA. Analytica Chimica Acta, 1122, 39–47.
Acknowledgements
The authors thank the financial and equipment support from The Third Affiliated Hospital of Chongqing Medical University.
Author information
Authors and Affiliations
Contributions
S.C. and G.M. designed the strategy, completed the preparation of the research, and wrote the manuscript; R.Y., Y.Z., S.D., and G.M. assisted data analysis.
Corresponding author
Ethics declarations
Ethical Approval
Permission from the Institutional Animal Ethical Committee was received before making these experiments.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 14.1 KB)
Rights and permissions
About this article
Cite this article
Sun, C., Rong, Y., Yang, Z. et al. Construction of Dual-Target Recognition-Based Specific MicroRNA Detection Method for Acute Pancreatitis Analysis. Appl Biochem Biotechnol 194, 3136–3144 (2022). https://doi.org/10.1007/s12010-022-03907-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-022-03907-7