Abstract
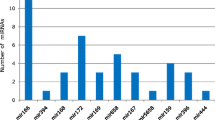
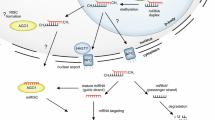
MicroRNAs are short, endogenous, non-coding RNAs, liable for essential regulatory function. Numerous miRNAs have been identified and studied in plants with known genomic or small RNA resources. Despite the availability of genomic and transcriptomic resources, the miRNAs have not been reported in the medicinal tree Azadirachta indica (Neem) till date. Here for the first time, we report extensive identification of miRNAs and their possible targets in A. indica which might help to unravel their therapeutic potential. A comprehensive search of miRNAs in the A. indica genome by C-mii tool was performed. Overall, 123 miRNAs classified into 63 families and their stem-loop hairpin structures were predicted. The size of the A. indica (ain)-miRNAs ranged between 19 and 23 nt in length, and their corresponding ain-miRNA precursor sequence MFEI value averaged as −1.147 kcal/mol. The targets of ain-miRNAs were predicted in A. indica as well as Arabidopsis thaliana plant. The gene ontology (GO) annotation revealed the involvement of ain-miRNA targets in developmental processes, transport, stress, and metabolic processes including secondary metabolism. Stem-loop qRT-PCR was carried out for 25 randomly selected ain-miRNAs and differential expression patterns were observed in different A. indica tissues. Expression of miRNAs and its targets shows negative correlation in a dependent manner.







Similar content being viewed by others
Abbreviations
- ain-miRNA:
-
Azadirachta indica microRNA
- DCL1:
-
Dicer-like-1
- EST:
-
expressed sequence tag
- GRFs :
-
growth regulating factors
- HEN1:
-
Hua-Enhancer1
- MFE:
-
minimum folding free energy
- MFEI:
-
minimal folding energy index
- NGS:
-
next-generation sequencing
- NF-YA :
-
nuclear transcription factor Y subunit alpha
- pri-miRNAs:
-
primary miRNA transcripts
- pre-miRNAs:
-
precursor miRNA transcripts
- RISC:
-
RNA-induced silencing complex
- SPL :
-
SQUAMOSA PROMOTER BINDING PROTEIN-LIKE
References
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297. https://doi.org/10.1016/S0092-8674(04)00045-5.
Kurihara, Y., & Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proceedings of the National Academy of Sciences, 101(34), 12753–12758. https://doi.org/10.1073/pnas.0403115101.
Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R. W., & Chen, X. (2005). Methylation as a crucial step in plant microRNA biogenesis. Science, 307(5711), 932–935. https://doi.org/10.1126/science.1107130.
Bollman, K. M., Aukerman, M. J., Park, M.-Y., Hunter, C., Berardini, T. Z., & Poethig, R. S. (2003). HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development, 130(8), 1493–1504. https://doi.org/10.1242/dev.00362.
Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell, 136(4), 669–687. https://doi.org/10.1016/j.cell.2009.01.046.
Jones-Rhoades, M. W., Bartel, D. P., & Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annual Reveiw Plant Biology, 57(1), 19–53. https://doi.org/10.1146/annurev.arplant.57.032905.105218.
Lee, R. C., Feinbaum, R. L., & Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 75(5), 843–854. https://doi.org/10.1016/0092-8674(93)90529-Y.
Feng, J., Wang, K., Liu, X., Chen, S., & Chen, J. (2009). The quantification of tomato microRNAs response to viral infection by stem-loop real-time RT-PCR. Gene, 437(1–2), 14–21. https://doi.org/10.1016/j.gene.2009.01.017.
Sunkar, R., Girke, T., Jain, P. K., & Zhu, J. K. (2005). Cloning and characterization of microRNAs from rice. The Plant Cell, 17(5), 1397–1411. https://doi.org/10.1105/tpc.105.031682.
Yao, Y., Guo, G., Ni, Z., Sunkar, R., Du, J., Zhu, J. K., & Sun, Q. (2007). Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biology, 8(6), R96.
Han, J., Li, A., Liu, H., Wen, X., Zhao, M., Korir, N. B., & Fang, J. (2014). Computational identification of microRNAs in the strawberry (Fragaria × ananassa) genome sequence and validation of their precise sequences by miR-RACE. Gene, 536(1), 151–162. https://doi.org/10.1016/j.gene.2013.11.023.
Han, J., Xie, H., Sun, Q., Wang, J., Lu, M., Wang, W., & Pan, J. (2014). Bioinformatic identification and experimental validation of miRNAs from foxtail millet (Setaria italica). Gene, 546(2), 367–377. https://doi.org/10.1016/j.gene.2014.05.050.
Abreu, P. M. V., Gaspar, C. G., Buss, D. S., Ventura, J. A., Ferreira, P. C. G., & Fernandes, P. M. B. (2014). Carica papaya microRNAs are responsive to Papaya meleira virus infection. PLoS One, 9(7), e103401. https://doi.org/10.1371/journal.pone.0103401.
Li, C., Zhu, Y., Guo, X., Sun, C., Luo, H., Song, J., & Chen, S. (2013). Transcriptome analysis reveals ginsenosides biosynthetic genes, microRNAs and simple sequence repeats in Panax ginseng C. A. Meyer. BMC Genomics, 14(1), 245.
Gébelin, V., Argout, X., Engchuan, W., Pitollat, B., Duan, C., Montoro, P., & Leclercq, J. (2012). Identification of novel microRNAs in Hevea brasiliensis and computational prediction of their targets. BMC Plant Biology, 12(1), 18. https://doi.org/10.1186/1471-2229-12-18.
Jain, M., Chevala, V. V. S. N., & Garg, R. (2014). Genome-wide discovery and differential regulation of conserved and novel microRNAs in chickpea via deep sequencing. Journal of Experimental Botany, 65(20), 5945–5958. https://doi.org/10.1093/jxb/eru333.
Prakash, P., Ghosliya, D., & Gupta, V. (2015). Identification of conserved and novel microRNAs in Catharanthus roseus by deep sequencing and computational prediction of their potential targets. Gene, 554(2), 181–195. https://doi.org/10.1016/j.gene.2014.10.046.
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., & Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell, 110(4), 513–520. https://doi.org/10.1016/S0092-8674(02)00863-2.
Khraiwesh, B., Zhu, J.-K., & Zhu, J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 1819(2), 137–148. https://doi.org/10.1016/j.bbagrm.2011.05.001.
Chuck, G., Candela, H., & Hake, S. (2009). Big impacts by small RNAs in plant development. Current Opinion in Plant Biology, 12(1), 81–86. https://doi.org/10.1016/j.pbi.2008.09.008.
Gou, J.-Y., Felippes, F. F., Liu, C. J., Weigel, D., & Wang, J.-W. (2011). Negative regulation of Anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. The Plant Cell, 23(4), 1512–1522. https://doi.org/10.1105/tpc.111.084525.
Sharma, D., Tiwari, M., Pandey, A., Bhatia, C., Sharma, A., & Trivedi, P. K. (2016). MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiology, 171(2), 944–959. https://doi.org/10.1104/pp.15.01831.
Ng, D. W. K., Zhang, C., Miller, M., Palmer, G., Whiteley, M., Tholl, D., & Chen, Z. J. (2011). Cis-and trans-Regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. The Plant Cell, 23(5), 1729–1740. https://doi.org/10.1105/tpc.111.083915.
Veitch, G. E., Boyer, A., & Ley, S. V. (2008). The azadirachtin story. Angewandte Chemie International Edition, 47(49), 9402–9429. https://doi.org/10.1002/anie.200802675.
Subramani, R., Gonzalez, E., Nandy, S. B., Arumugam, A., Camacho, F., Medel, J., & Lakshmanaswamy, R. (2017). Gedunin inhibits pancreatic cancer by altering sonic hedgehog signaling pathway. Oncotarget, 8(7), 10891. https://dx.doi.org/10.18632%2Foncotarget.8055–10904.
Ponnusamy, S., Haldar, S., Mulani, F., Zinjarde, S., Thulasiram, H., & RaviKumar, A. (2015). Gedunin and azadiradione: human pancreatic alpha-amylase inhibiting limonoids from neem (Azadirachta indica) as anti-diabetic agents. PLoS One, 10(10), e0140113. https://doi.org/10.1371/journal.pone.0140113.
Wang, L., Do Dang Khoa Phan, J. Z., Ong, P.-S., Thuya, W. L., Soo, R., Wong, A. L. A., & Sethi, G. (2016). Anticancer properties of nimbolide and pharmacokinetic considerations to accelerate its development. Oncotarget, 7(28), 44790. https://dx.doi.org/10.18632%2Foncotarget.8316–44802.
Biswas, K., Chattopadhyay, I., Banerjee, R. K., & Bandyopadhyay, U. (2002). Biological activities and medicinal properties of neem (Azadirachta indica). Current Science, 82(11), 1336–1345.
Brahmachari, G. (2004). Neem–an omnipotent plant: a retrospection. Chembiochem, 5(4), 408–421. https://doi.org/10.1002/cbic.200300749.
Krishnan, N. M., Pattnaik, S., Deepak, S. A., Hariharan, A. K., Gaur, P., Chaudhary, R., & Panda, B. (2011). De novo sequencing and assembly of Azadirachta indica fruit transcriptome. Current Science, 1553–1561.
Krishnan, N. M., Jain, P., Gupta, S., Hariharan, A. K., & Panda, B. (2016). An Improved Genome Assembly of Azadirachta indica A. Juss. G3: Genes, Genomes, Genetics, 6(7), 1835–1840. https://doi.org/10.1534/g3.116.030056.
Krishnan, N. M., Pattnaik, S., Jain, P., Gaur, P., Choudhary, R., Vaidyanathan, S., & Nair, J. (2012). A draft of the genome and four transcriptomes of a medicinal and pesticidal angiosperm Azadirachta indica. BMC Genomics, 13(1), 1–13.
Rajakani, R., Narnoliya, L., Sangwan, N. S., Sangwan, R. S., & Gupta, V. (2014). Subtractive transcriptomes of fruit and leaf reveal differential representation of transcripts in Azadirachta indica. Tree Genetics & Genomes, 10(5), 1331–1351. https://doi.org/10.1007/s11295-014-0764-7.
Narnoliya, L. K., Rajakani, R., Sangwan, N. S., Gupta, V., & Sangwan, R. S. (2014). Comparative transcripts profiling of fruit mesocarp and endocarp relevant to secondary metabolism by suppression subtractive hybridization in Azadirachta indica (neem). Molecular Biology Reports, 41(5), 3147–3162. https://doi.org/10.1007/s11033-014-3174-x.
Kuravadi, N. A., Yenagi, V., Rangiah, K., Mahesh, H. B., Rajamani, A., Shirke, M. D., & Siddappa, S. (2015). Comprehensive analyses of genomes, transcriptomes and metabolites of neem tree. PeerJ, 3, e1066. https://doi.org/10.7717/peerj.1066.
Pandreka, A., Dandekar, D. S., Haldar, S., Uttara, V., Vijayshree, S. G., Mulani, F. A., & Thulasiram, H. V. (2015). Triterpenoid profiling and functional characterization of the initial genes involved in isoprenoid biosynthesis in neem (Azadirachta indica). BMC Plant Biology, 15(1), 214. https://doi.org/10.1186/s12870-015-0593-3.
Numnark, S., Mhuantong, W., Ingsriswang, S., & Wichadakul, D. (2012). C-mii: a tool for plant miRNA and target identification. BMC Genomics, 13(Suppl 7), S16. https://doi.org/10.1186/1471-2164-13-S7-S16.
Meyers, B. C., Axtell, M. J., Bartel, B., Bartel, D. P., Baulcombe, D., Bowman, J. L., & Green, P. J. (2008). Criteria for annotation of plant MicroRNAs. The Plant Cell, 20(12), 3186–3190. https://doi.org/10.1105/tpc.108.064311.
Prakash, P., Rajakani, R., & Gupta, V. (2016). Transcriptome-wide identification of Rauvolfia serpentina microRNAs and prediction of their potential targets. Computational Biology and Chemistry, 61, 62–74. https://doi.org/10.1016/j.compbiolchem.2015.12.002.
Dai, X., & Zhao, P. X. (2011). psRNA Target: a plant small RNA target analysis server. Nucleic Acids Research, 39(Suppl 2), W155–W159. https://doi.org/10.1093/nar/gkr319.
Conesa, A., Gotz, S., Miguel, J., Gomez, G., Terol, J., Talon, M., & Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21(18), 3674–3676. https://doi.org/10.1093/bioinformatics/bti610.
Rajakani, R., Narnoliya, L., Sangwan, N. S., Sangwan, R. S., & Gupta, V. (2013). Activated charcoal-mediated RNA extraction method for Azadirachta indica and plants highly rich in polyphenolics, polysaccharides and other complex secondary compounds. BMC Research Notes, 6(1), 125. https://doi.org/10.1186/1756-0500-6-125.
Varkonyi-Gasic, E., Wu, R., Wood, M., Walton, E. F., & Hellens, R. P. (2007). Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods, 3(1), 1–12. https://doi.org/10.1186/1746-4811-3-12.
Schmittgen, T. D., & Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C T method. Nature Protocols, 3(6), 1101–1108. https://doi.org/10.1038/nprot.2008.73.
Archak, S., & Nagaraju, J. (2007). Computational prediction of rice (Oryza sativa) miRNA targets. Genomics, Proteomics & Bioinformatics, 5(3–4), 196–206. https://doi.org/10.1016/S1672-0229(08)60007-8.
He, C., Li, Y.-X., Zhang, G., Gu, Z., Yang, R., Li, J., & Wang, J. (2012). MiRmat: mature microRNA sequence prediction. PLoS One, 7(12), e51673. https://doi.org/10.1371/journal.pone.0051673.
Wu, Y., Wei, B., Liu, H., Li, T., & Rayner, S. (2011). MiRPara: a SVM-based software tool for prediction of most probable microRNA coding regions in genome scale sequences. BMC Bioinformatics, 12(1), 1–14.
Tyagi, S., Vaz, C., Gupta, V., Bhatia, R., Maheshwari, S., Srinivasan, A., & Bhattacharya, A. (2008). CID-miRNA: a web server for prediction of novel miRNA precursors in human genome. Biochemical and Biophysical Research Communications, 372(4), 831–834. https://doi.org/10.1016/j.bbrc.2008.05.134.
Grundhoff, A., Sullivan, C. S., & Ganem, D. (2006). A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpes viruses. RNA, 12(5), 733–750. https://doi.org/10.1261/rna.2326106.
Meng, J., Liu, D., Sun, C., & Luan, Y. (2014). Prediction of plant pre-microRNAs and their microRNAs in genome-scale sequences using structure-sequence features and support vector machine. BMC Bioinformatics, 15(1), 423. https://doi.org/10.1186/s12859-014-0423-x.
Mendes, N. D., Freitas, A. T., & Sagot, M.-F. (2009). Current tools for the identification of miRNA genes and their targets. Nucleic Acids Research, 37(8), 2419–2433. https://doi.org/10.1093/nar/gkp145.
Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B., & Bartel, D. P. (2002). MicroRNAs in plants. Genes and Development, 16(13), 1616–1626. https://doi.org/10.1101/gad.1004402.
Chi, X., Yang, Q., Chen, X., Wang, J., Pan, L., Chen, M., & Yu, S. (2011). Identification and characterization of microRNAs from peanut (Arachis hypogaea L.) by high-throughput sequencing. PLoS One, 6(11), e27530. https://doi.org/10.1371/journal.pone.0027530.
Xie, K., Wu, C., & Xiong, L. (2006). Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiology, 142(1), 280–293. https://doi.org/10.1104/pp.106.084475.
He, J., Xu, M., Willmann, M. R., McCormick, K., Hu, T., Yang, L., & Poethig, R. S. (2018). Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana. PLoS Genetics, 14(4), e1007337. https://doi.org/10.1371/journal.pgen.1007337.
Wu, G., Park, M. Y., Conway, S. R., Wang, J. W., Weigel, D., & Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell, 138(4), 750–759. https://doi.org/10.1016/j.cell.2009.06.031.
Xu, M. Y., Zhu, J. X., Zhang, M., & Wang, L. (2016). Advances on plant miR169/NF-YA regulation modules. Yi Chuan = Hereditas, 38(8), 700–706. https://doi.org/10.16288/j.yczz.15-526.
Casadevall, R., Rodriguez, R. E., Debernardi, J. M., Palatnik, J. F., & Casati, P. (2013). Repression of growth regulating factors by the microRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. The Plant Cell, 25(9), 3570–3583. https://doi.org/10.1105/tpc.113.117473.
Bari, R., Pant, B. D., Stitt, M., & Scheible, W. R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiology, 141(3), 988–999. https://doi.org/10.1104/pp.106.079707.
Yue, E., Liu, Z., Li, C., Li, Y., Liu, Q., & Xu, J.-H. (2017). Overexpression of miR529a confers enhanced resistance to oxidative stress in rice (Oryza sativa L.). Plant Cell Reports, 36(7), 1171–1182. https://doi.org/10.1007/s00299-017-2146-8.
Ren, J., Zhou, J. J., Duan, W. K., Song, X. M., Liu, T. K., Hou, X. L., & Li, Y. (2014). Copper stress induces the differential expression of microRNAs in non-heading Chinese cabbage. Biologia Plantarum, 58(3), 491–498. https://doi.org/10.1007/s10535-014-0426-5.
Pei, H., Ma, N., Chen, J., Zheng, Y., Tian, J., Li, J., & Gao, J. (2013). Integrative analysis of miRNA and mRNA profiles in response to ethylene in rose petals during flower opening. PLoS One, 8(5), e64290. https://doi.org/10.1371/journal.pone.0064290.
Acknowledgments
This research work was financially supported under BSC0203 and Institutional major laboratory project (MLP04) by Council of Scientific and Industrial Research (CSIR), Govt. of India. Research fellowship from University Grants Commission, Govt. of India, is duly acknowledged. AcSIR is acknowledged for the academic support. The authors are sincerely thankful to the Director, CSIR-CIMAP, for encouragement, support and providing the required laboratory facilities.
Author information
Authors and Affiliations
Contributions
RR and VG conceived and designed the experimental plans. RR, PP, and DG applied the plan and carried out experiments. RS and AS helped in data collection and analysis of experiments. RR carried out the stem-loop qRT-PCR, qPCR analysis, and wrote the manuscript. VG guided the experimental designs and finalized the manuscript. All the authors have read and agree to the content of the finalized manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Fig. 1
Schematic representation of in silico analysis strategy used for the identification of ain-miRNAs and their potential mRNA transcript targets (PPTX 82 kb)
Supplementary Fig. 2
Stem-loop hairpin structures of C-mii predicted 123 A. indica miRNAs. Mature ain-miRNA nucleotides are marked in red. The precursor’s nucleotide positions in primary ain-miRNAs are indicated in brackets (PPTX 1878 kb)
Supplementary Figure 2
(PDF 911 kb)
Supplementary Fig. 3
Nucleotide dominance in each base position and nucleotide probability analysis in identified mature ain-miRNA sequences (PPTX 512 kb)
Supplementary Fig. 4
a Blast2GO examination of microRNA targets predicted in A. indica leaf transcriptome SRX096301, b Blast2GO analysis of miRNA targets predicted in A. indica seed transcriptome SRR342216 (PPTX 388 kb)
Supplementary Fig. 5
Expression analyses of ain-miRNAs in different neem tissues (immature fruit, leaf, fruit mesocarp, and fruit endocarp) by using semi-quantitative stem-loop RT-PCR. Randomly selected ain-miRNAs were checked for transcript/expression level (miR156, miR169g, miR169h, miR172, miR396b, miR399, miR403c, miR5021, miR5139, miR5522, miR1533f, miR1533g, miR5512, miR157, miR529, miR776, miR1088-5p, miR1511, miR1525, miR5025, miR1533a, miR1533c, miR1533i, miR1533d, and miR1533h) (XLSX 13 kb)
Supplementary Table 1
Primers used in stem-loop qRT-PCR and qRT-PCR studies (XLS 70 kb)
Supplementary Table 2
A. indica mature miRNA sequences with number of mismatches between miRNA and miRNA*, primary miRNA MFE (kcal/mol), precursor miRNA MFE, and precursor miRNA MFEI (kcal/mol) (DOCX 33 kb)
Supplementary Table 3
A. indica mature miRNA sequences identified by C-mii-based computer prediction and their characteristic features (XLS 69 kb)
Supplementary Table 4
psRNATarget analysis of miRNA targets predicted in A. thaliana transcriptome by using the identified A. indica mature miRNA sequences as query (XLS 114 kb)
Supplementary Table 5
Blast2GO analysis of miRNA targets predicted in A. indica leaf transcriptome SRX096301 by using the identified A. indica mature miRNA sequences as query (XLS 63 kb)
Supplementary Table 6
Blast2GO analysis of miRNA targets predicted in A. indica seed transcriptome SRR342216 by using the identified A. indica mature miRNA sequences as query (XLS 63 kb)
Rights and permissions
About this article
Cite this article
Rajakani, R., Prakash, P., Ghosliya, D. et al. Azadirachta indica MicroRNAs: Genome-Wide Identification, Target Transcript Prediction, and Expression Analyses. Appl Biochem Biotechnol 193, 1924–1944 (2021). https://doi.org/10.1007/s12010-021-03500-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03500-4