Abstract
Isomaltooligosaccharides (IMO), the glucosylsaccharides used as food additives, are made from saccharified starch by enzymes or microbial cells with transglycosylation activity. This study aimed to generate shuffled futants of Aspergillus niger with enhanced transglycosylation activity for industrial IMO production. The starting mutant population was generated by 60Co-γ radiation; mutants with higher transglycosylation activity were selected and subjected to recursive protoplast fusion. The resulting fusants were screened by a novel high-throughput method based on detecting non-fermentable reducing sugar. After three rounds of genome shuffling, the best performing strain GS3-3 was obtained, its transglycosylation activity (14.91 U/mL) was increased by 194.1 % compared to that of original strain C-6181. In fermentor test, transglycosylation activity of GS3-3 was obtained at 16.61 U/mL. The mycelia of GS3-3 were reused ten times to produce IMO syrup from liquefied cassava starch containing about 280 g/L total sugar within 4 days. The conversion of liquefied cassava starch to IMO was at 71.3–72.1 %, which was higher than the best conversion (68 %) ever reported. GS3-3 shows a great potential for industrial IMO production.
Similar content being viewed by others
Introduction
Isomaltooligosaccharides (IMO) are glucosylsaccharides with one or more α-1,6-glucosidic linkages, including isomaltose, panose, isomaltotriose, and other branched oligosaccharides. Since 1980s, they have attracted many attentions and have been widely used as food additives due to their many unique features. They can effectively promote the growth and reproduction of probiotic Bifidobacterium, prevent dental caries and aging of starch, and prolong food preservation time [1].
Traditionally, IMO are converted from saccharified starch by enzymes or microbial cells with transglycosylation activity [2, 3]. Enzymes, such as d-glucosyltransferase (EC2.4.1.24) and neopullulanase (EC 3.2.1.135), and microbial cells with transglycosylation activity catalyze both hydrolytic and transfer reactions of α-d-gluco-oligosaccharides and produce a class of α-(1, 6)-linked oligomers of glucose, such as IMO. Enhancing transglycosylation activity of these catalysts can improve the efficiency of IMO production. Many investigators have exerted their efforts to improve transglycosylation activity of enzymes by various methods, including purification and characterization of newly found enzymes [4], heterologous expression of enzymes with transglycosylation activity [5], immobilization of the enzymes [6], etc. In contrast to enzymes, few studies on whole cell transglycosylation biocatalyst have been published in the past decade.
During the past several years, an efficient technology named genome shuffling had been used to improve industrially important microbial phenotypes. Genome shuffling is the recombination between multiple parents of each generation, and several rounds of genome fusion are carried out. The advantage of this technique is that the genetic breeding can be performed on the tested microbes without knowing their genetic background, making it a highly effective method. Recent reports have described the use of genome shuffling to improve production of low-temperature alkaliphilic lipase by Acinetobacter johnsonii [7], cellulase by Penicillium decumbens JU-A10 [8], and xylanase by Aspergillus sp. NRCF5 [9]. However, little work has been done on improvement of transglycosylation activity through genome shuffling technology, probably due to the lack of an efficient high-throughput method [10], indicating the need to develop new high-throughput methods that can be used to increase the screening efficiency for a large number of fusion strains.
Transglycosylation activity was normally determined by using high-performance liquid chromatography (HPLC) [11–13]. Although HPLC is intuitive and accurate, its high cost and low efficiency hinder its application in large-scale screening of mutants. Our previous work established a novel high-throughput method that can rapidly screen a large number of mutants with high transglycosylation activity in Aspergillus niger [14]. A. niger has been used already for many decades to produce food enzymes and organic acids and is recognized as a relatively safe production organism by FAO/WHO experts [15]. Therefore, A. niger strain with high transglycosylation activity may have potential for industrial IMO production. In the present study, we combined genome shuffling with the high-throughput method to rapidly and efficiently generate high-yield recombinant with high transglycosylation activity in A. niger.
Materials and Methods
Microorganisms and Mutation
Strain C-6181 with transglycosylation activity was isolated from mutants of A. niger GXM-2 and preserved in our laboratory. Mycelium was grown at 37 °C for 48 h on solid medium (malt extract 20.0 g/L, glucose 20.0 g/L, and agar 20.0 g/L; pH 5.5). Spores were collected with 0.85 % (w/v) sterile saline and prepared as 1 × 108 spores/mL suspension. The spore suspension was irradiated with 60Co-γ radiation of 1,500 Gy to get a survival rate of 11 %. (The irradiation services were provided by Guangxi Shenzhou Irradiation Center Co., Ltd., Nanning, China). The irradiated spore suspension was then diluted and spread on trypan blue-starch medium (soluble starch 10.0 g/L, MgSO4 0.5 g/L, KCl 0.5 g/L, FeSO4 0.05 g/L, K2HPO4 0.5 g/L, NaNO3 3.0 g/L, NaCl 70.0 g/L, trypan blue 0.2 g/L, and agar 15.0 g/L; pH 6.0) and incubated at 37 °C for 48 h. Mutants were screened by a previously described high-throughput method [14]. Those mutants showing higher transglycosylation activity were selected as the starters for genome shuffling.
Protoplast Preparation and Regeneration
Protoplasts were formed by a previously reported method with modification [16–18]. Each mutant strain was inoculated in 50 mL of protoplast medium (sucrose 30.0 g/L, NaNO3 3.0 g/L, yeast extract 5.0 g/L, tryptone 5.0 g/L, K2HPO4 1.0 g/L, MgSO4 0.5 g/L, KCl 0.5 g/L, and FeSO4 0.01 g/L; pH 6.0) with a stock spore suspension (2 %, v/v, 106 spores/mL suspension) and incubated at 37 °C for 10 h on a rotary shaker (180 rpm). The mycelia were harvested by filtration over a Büchner funnel with nylon gauze, washed twice with sterile distilled water, resuspended in 5 mL of citrate phosphate (CP) buffer (0.1 M, pH 5.8), containing 0.7 M NaCl and 0.2 M CaCl2 dissolved with enzyme cocktail containing 2 mg/mL of cellulase (CAS 9012-54-8, Sigma-Aldrich, St. Louis, MO, USA), 3 mg/mL of snail enzyme (Fujian Zhangzhou Jintian Bio-tech Co., Ltd., Fujiang, China), and 1 mg/mL of lytic enzyme (CAS 158928, Qiagen, Germantown, MD, USA)), and incubated with gentle shaking for up to 5 h. The released protoplasts were photographed using a phase-contrast microscope. Mycelial debris were removed by filtration through 10 μm nylon mesh. Protoplasts were washed twice in 8 mL CP buffer after centrifugation at 2,000 rpm at 4 °C for 10 min. The rates of protoplast preparation and regeneration were obtained by determining colony counts using the following formulas:
Where A represents the total number of colonies counted on trypan blue-starch medium before hydrolysis of cell wall by enzyme cocktail, B represents the number of colonies counted on trypan blue-starch medium containing 0.7 M NaCl after hydrolysis of cell wall, and C represents the number of colonies counted on trypan blue-starch medium after hydrolysis of cell wall.
Protoplast Inactivation
The protoplast suspensions were inactivated by heat or UV irradiation. For heat treatment, the protoplast suspensions were incubated in 60 °C water bath for 5, 10, 12.5, 15, 17.5, and 20 min to choose the optimal inactivation condition. For UV treatment, the protoplast suspensions were placed under a preheated 30 W UV lamp at a vertical distance of 20 cm and irradiated for 10, 20, 30, 40, 45, and 50 min to choose the optimal inactivation condition. The treated protoplasts were maintained in the dark for 5 h to avoid photoreactivation repair. Inactivation was confirmed by lack of growth on the trypan blue-starch medium containing 0.7 M NaCl at 37 °C.
Genome Shuffling
Genome shuffling was carried out using the described methods with modifications [3, 16–18]. Equal number of protoplasts from different parental populations were mixed and divided equally into two parts. One part was inactivated with UV irradiation for 20 min and the other was heated at 60 °C for 40 min. Both inactivated protoplasts were mixed in a 1:1 cell ratio, centrifuged, and resuspended in 0.5 ml PB buffer (20 mM Na2HPO4-NaH2PO4, pH 6) supplemented with 0.7 M NaCl and 0.2 M CaCl2. Then, 4.5 ml of PB buffer containing 30 % (w/v) polyethylene glycol 6000 was added, and the mixture was gently shaken at 37 °C for 10 min. Then, the suspension was diluted fivefold. The fused protoplasts were centrifuged, washed twice with PB buffer, and resuspended in 5 mL of PB buffer. It was serially 10 × diluted and spread on trypan blue-starch medium containing 0.7 M NaCl and incubated at 37 °C for 60 h. Subsequent rounds of genome shuffling were carried out by repeating the protoplast fusion described above.
Screening of Fusants
The fusants were screened by the high-throughput method [14]. The schematic flow chart of screening fusants was shown in Fig. 1.
Determination of Transglucosidase Activity
Five milliliter of spore suspension (106 spores/mL) of each possible strain with enhanced transglycosylation activity was inoculated to a 250 mL flask containing 45 mL of fermentation medium B (liquefied cassava starch 100 g/L and malt extract 20 g/L, pH 5.5) and cultured at 37 °C, 160 rpm for 60 h. Mycelia were harvested by filtration and washed twice with deionized water. Crude enzyme extract was prepared, and their transglycosylation activity was assayed as previously reported [13]. HPLC analysis was performed on a Kromasil NH2 column (5 μm, 4.6 mm × 250 mm) under the conditions of mobile phase of acetonitrile/water (75:25, v/v) and a flow rate of 1.0 mL/min. One micromole pentose produced by the enzyme at 37 °C and pH 5.0 per hour was defined as one activity unit.
Fermentor Cultivation
A 3-L fermentor (INFORS-HT Labfors 3, Switzerland) containing 1,800 mL of fermentation medium B was inoculated with 210 mL of 24-h-old seed culture. The process was controlled at 37 °C, pH 5.5, 400 rpm agitation speed, and 0.35 vvm aeration for 48 h. Biomass, pH, residual sugar, and transglycosylation activity were monitored during fermentation, according to a fixed time schedule, at time zero and after 8, 16, 24, 32, 40, and 48 h. Eighty milliliter of culture medium was collected from each sample. Mycelia were harvested by filtration and washed twice with deionized water.
Preparation of Liquefied Cassava Starch
Cassava starch slurry (30 %, w/v) was prepared and dissolved in 2 L of deionized water and liquefied by Termamyl SC (1 g/kg of cassava starch) at 85–90 °C for 40–60 min. Liquefaction was stopped according to the iodine test for starch. The turning of color to red indicates the complete conversion of starch to dextrin. Then all of the mixture was transferred to a 3 L stirred bioreactor. The amount of water that was lost during heating was made up, and the temperature was raised gradually to 121 °C for over 15 min. Termamyl SC was inactivated with sterilization.
Isomaltooligosaccharides Production and Determination
The resultant liquefied mixture was incubated with Fungamyl 800 L (1 g/kg of cassava starch) and the mycelia described above in stirred bioreactor at 55 °C with agitation rates of 50 rpm. Produced syrup was separated from mycelia by filtration and heated at 90 °C for 5 min to inactivate enzymes. Mycelia were harvested and then reused to next batch. IMO concentration in syrup was assayed by HPLC using the same condition as described above in “determination of transglucosidase activity”. Glucose, maltose and maltotriose (CAS 1109-28-0), isomaltose (CAS 499-40-1), panose (CAS 33401-87-5), isomaltotriose (CAS 3371-50-4), and isomaltotriose (CAS 35997-20-7) (all were purchased from Sigma-Aldrich) were used as standards. Their ratios of peak areas to concentrations were used to convert peak areas of sugars found in the products into concentration. In order to facilitate detection, all DP4 branched oligosaccharides are accounted for isomaltotetraose. Total sugar concentration was assayed with enthrone reagent according to the Scott and Melvin procedure [19]. IMO yield was calculated using the following formula:
Where A represents the total sugar concentration of product and B represents the total IMO concentration of product.
Results
Strain Mutagenesis and Mutant Screening
Genome shuffling accelerates directed evolution by facilitating recombination within a diverse mutant population. Thus, utilization of this method requires a diverse population of mutants with an improvement of the desired phenotype compared with the original strain as the starting point [20, 21]. To fulfill this need, we used 60Co-γ radiation as the mutagenic agent to introduce the genetic variability.
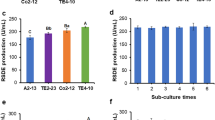
Mutants with the maximum transglycosylation activity were selected from a mutant library consisting of over 3,000 mutants. Forty-two mutants showing higher non-fermentable reducing sugar production were picked out for shake flask fermentation test, and their crude enzyme extracts were prepared to determine the transglycosylation activity. Five mutants, namely CM-115, CM-798, CM-1357, CM-1526, and CM-2761, exhibited the higher transglycosylation activity in their crude enzyme extracts with 5.45, 5.76, 5.39, 5.81, and 5.52 U/mL, respectively, than that of the original strains (4.77 U/mL; Fig. 1). These mutants were adopted as the starting population for genome shuffling.
Preparation and Regeneration of Protoplasts
Because A. niger C-6181 was the parent strain of the resulting mutants, it was used in the tests for a series of single factors to optimize protoplast preparation and regeneration condition before protoplast fusion was performed. The optimal conditions were described under materials and methods. The optimal rates of protoplast preparation and regeneration were over 93 and 55 %, respectively.
Genome Shuffling
The conditions of inactivation process reported in previous studies [2, 22, 23] varied significantly. In this study, we observed that harsh conditions of inactivation process led the significant decreases in survival rate, making the cells more difficult to resuscitate. Alternatively, a relatively mild condition led to a high survival ratio among the starting strains, a situation that resulted in difficulty in identifying the properties of the strains cultured on the resurrection plates. Because of this problem, we optimized the condition for inactivation process of A. niger protoplasts and observed that incubation of protoplasts in 60 °C water bath for 20 min resulted in a 100 % lethal rate, whereas irradiation of protoplasts with UV inactivation for 40 min resulted in a 100 % lethal rate.
Genome shuffling is dependent upon the recursive fusion of protoplasts to allow recombination. This recursive strategy permits quick acquisition of the interested phenotypes. The efficiency of genome shuffling is related to the efficiencies of formation, fusion, and regeneration of protoplasts [24, 25]. The optimal protoplast fusion condition described under materials and methods were achieved using single factor tests.
Five initial mutants, namely CM-115, CM-798, CM-1357, CM-1526, and CM-2761, were subjected to the first round of pool-wise recursive protoplast fusion. After the first fusion, 500 rapidly growing colonies with large transparent circles were picked out from the trypan blue-starch plates for fermentation test in 50 mL sterile Eppendorf tubes, and 18 strains were selected as higher non-fermentable reducing sugar producers. Among them, four strains showed the highest transglycosylation activity from 7.63 to 8.11 U/mL, which was increased by 50–59.9 % as compared with that of strain C-6181(5.07 U/mL). They were then adopted for the second round of shuffling.
Five out of 26 strains showing higher non-fermentable reducing sugar production were selected from the second shuffled library. Their transglycosylation activity reached 10.78–11.42 U/mL (Fig. 2). These strains were subjected to the third round of shuffling.
Comparison of transglycosylation activity between original strain (C-6181), mutants (CM) and fusants (GS) in shake flask fermentation tests. GS1, GS2, and GS3 strains were isolated from the first, the second, and the third rounds of genome shuffling, respectively. All the data represent mean values of three independent experiments and the error bars represent the standard deviations
After the third round shuffling, three isolates, namely GS3-1, GS3-2, and GS3-3, respectively, with higher non-fermentable reducing sugar production than the starting strains of this round were screened from 19 strains. GS3-1, GS3-2, and GS3-3 showed significantly higher transglycosylation activity, reaching 14.78, 14.62, and 14.91 U/mL, respectively.
Comparison of C-6181 and GS3-3 in Bioreactor Fermentation
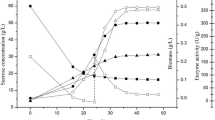
The performance of GS3-3 was compared with that of C-6181 in a 3 L fermentor. There were no significant differences in final pH and biomass between two strains. However, the transglycosylation activity accumulated more rapidly in GS3-3, its peak reached 16.61 U/mL, which was about 213 % higher than that of original strain (Fig. 3). Moreover, GS3-3 exhibited more rapid sugar consumption and completed the batch fermentation at 52 h, whereas strain C-6181 needed 72 h to finish it. The results showed that more IMO can be produced by the same number of GS3-3 cells within equal time compared with that of C-6181.
Production of Isomaltooligosaccharides
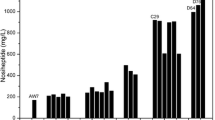
In previous study [14], we found that the mycelia of A. niger C-6181 can be used as whole cell biocatalyst to convert liquefied cassava starch to IMO syrup. Thus, IMO productivity of GS3-3 was tested. Its mycelia were reutilized to convert liquefied cassava starch to IMO syrup by ten times within 4 days. The optimal conversion time determined in a preliminary test was 8 h. As shown in Fig. 4, the changes in total IMO concentrations and the sum of isomaltose, panose, and isomaltotriose of the final products were relatively stable and achieved as high as 200.1–206.6 and 116.6–124.7 g/L, respectively. Further details of the final products were shown in Table 1.
a Time course of IMO (containing isomaltose, panose, isomaltotriose, and all DP4 branched oligosaccharides) production and total sugar consumption with A. niger GS3-3 in 3 L stirred bioreactor. b Time course of isomaltose, panose, and isomaltotriose production by A. niger GS3-3. All the data represent mean values of three independent experiments and the error bars represent the standard deviations
Discussion
Several enzymes and microbial cells are capable of catalyzing both the hydrolytic and transfer reactions of α-d-gluco-oligosaccharides in the IMO production. A. niger strains which have high transglycosylation activity will be the potentially important microorganism for industrial IMO production. The full genome sequence of A. niger CBS 513.88 was determined and annotated in 2007 [26]. A detailed full genome search showed the presence of a large number of previously unknown predicted enzymes belonging to the α-amylase superfamily, and many of them showed transglycosylation activity [22]. Therefore, it is difficult to explain the details of transglycosylation activity and its underlying regulatory mechanism in A. niger cells. In a previous study [14], we found that A. niger mutant C-6181 can convert liquefied cassava starch to high purity of IMO syrup, accounting for 83.7 % (w/w) of total sugar in product. Unfortunately, the conversion time was still too long for industrial IMO production. Consequently, to improve the transglycosylation activity of A. niger, C-6181 is needed, even though the cause of this high transglycosylation activity remains unclear.
Although traditional breeding methods have succeeded in generating many industrial strains [27, 28], it is a time-consuming and low-efficiency process. Genome shuffling is a powerful means for rapid breeding of improved organisms. It is a technique that allows for the recombination of several genomes simultaneously at different sites without the necessity for knowing the detailed genomic information. Therefore, multiple exchanges and multiple-gene recombination can occur rapidly and efficiently, resulting in the generation of a large number of strains that can then be tested for the desired phenotype [29]. Use of the un-inactivated parental cells for gene recombination could seriously interfere with the effective screening of fusants, due to the presence of a large number of unfused cells that can survive. Genome shuffling is much easier to be carried out by using inactivated parental protoplasts than viable ones [29]. Moreover, if parental protoplasts are inactivated by only one method, it may also be difficult to regenerate them [30]. Therefore in this study, according to the principle of complementary protoplast damage, we tested two different methods of inactivating parent protoplast prior to carrying out the fusion procedure. We observed that this approach increased the screening efficiency of the recombinants. Recently, several novel methods have been applied to improve the efficacy of recombinants screening, such as the application of genetic markers [31] and the addition of analog of product [30]. However, inactivated parental protoplast fusion is much less expensive and more convenient to operate. Therefore, it was applied in our study. Genome shuffling is applicable to the improvement of most complex phenotypes of microorganism [32] like the transglycosylation activity of A. niger C-6181. Moreover, development of a high-throughput screening method is the crucial step to ensure the success of the whole procedure of genome shuffling [30]. In this report, a shuffled strain GS3-3 adopting the same method produced a relatively high transglycosylation activity (14.91 U/mL) than all of ever reported.
The conventional method for the production of IMO from starch includes first degradation of starch into α-(1, 4)-linked α-d-gluco-oligosaccharides using α-amylase (EC.3.2.1.1), pullulanase (EC3.2.1.41), and β-amylase (EC3.2.1.2) and then conversion to α-(1, 6)-linked oligosaccharides using the enzymes with transglycosylation activity such as d-glucosyltransferase (EC2.4.1.24), (EC3.2.1.20), and neopullulanase (EC3.2.1.135). Unfortunately, all of these enzymes can be used only one time in the reactions [33]. To find alternative ways, many studies have been carried out to produce IMO using immobilized enzymes and permeabilized cells [6, 34, 35]. Besides starch, maltose syrup can be used to produce IMO as well [34]. However, according to the published reports for the production of IMO, whatever enzyme or cell is used as the biocatalyst, the highest purity of IMO is still at about 60 % (w/w), with glucose and α-(1, 4)-linked oligomers such as maltose as the by-products. Recently, the purity of IMO has been increased to 68 % by using the cooperative action of maltogenic amylase and α-glucantransferase (EC 2.4.1.25) [36]. In the present study, we were capable of converting the liquefied cassava starch to IMO by repeated batch reactions using mycelia of A. niger GS3-3 without permeabilized and immobilized steps. Furthermore, the mycelia were recycled by ten times and still produce IMO with good yield (71.3–72.1 %, w/w), which is much higher than the yield of 47.3–48.4 % reported previously [34, 37, 38] and slightly higher than the maximum yield at 68 % (w/w) reported by Lee et al. [36], indicating a good improvement in microbial cell-catalyzed IMO production.
In conclusion, the success of novel genome shuffling depends on the selection of initial variants, the efficiency of the genetic shuffling during meiosis and conjugation, and the power of the selection approaches. Through three consecutive rounds of genome shuffling in combination with an efficient screening by using a novel high throughput method, we have successfully selected the shuffled strain, GS3-3, that exhibited significantly higher transglycosylation activity as compared with that of the original strain, C-6181. More importantly, the mycelia of GS3-3 can be reused several times as whole cell biocatalyst to produce IMO from liquefied cassava starch with relatively high purity. The novel process is simple, efficient, and costs much less when compared to traditional methods of IMO production using either enzymes or microbial cells. To our knowledge, this is the first study to improve the transglycosylation activity of A. niger cells through genome shuffling. Our results should contribute to improving the efficiency of industrial IMO production.
References
Møller, M. S., Fredslund, F., Majumder, A., Nakai, H., Poulsen, J., Lo, C. N., Leggio, L., Svenson, B., & Abou Achem, M. (2012). Journal of Bacteriology, 194(16), 4249–4259.
Kuriki, T., Yanase, M., Takata, H., Takesada, Y., Imanaka, T., & Okada, S. (1993). Applied and Environmental Microbiology, 59, 953–959.
Khattab, A. A., & Bazaraa, W. A. (2005). Journal of Industrial Microbiology and Biotechnology, 32, 289–294.
Suzuki, R., Terasawa, K., Kimura, K., Fujimoto, Z., Momma, M., Kobayashi, M., Kimura, A., & Funane, K. (2012). Biochimica Et Biophysica Acta-Proteins and Proteomics, 1824(7), 919–924.
Chen, D. L., Tong, X., Chen, S. W., Chen, S., Wu, D., Fang, S. G., Wu, J., & Chen, J. (2010). Journal of Agricultural and Food Chemistry, 58(8), 4819–4824.
Mendis, M., Mendoza, B. R., & Simsek, S. (2012). Catalysis Letters, 142(9), 1107–1113.
Wang, H., Zhang, J., Wang, X., Qi, W., & Dai, Y. (2012). Biotechnology Letters, 34, 145–151.
Cheng, Y., Song, X., Qin, Y., & Qu, Y. (2009). Journal of Applied Microbiology, 107, 1837–1846.
El-Bondkly, A. M. (2012). Applied Biochemistry and Biotechnology, 167, 2160–2173.
Persson, M., & Palcic, M. M. (2008). Analytical Biochemistry, 378, 1–7.
Bousquet, M. P., Willemot, R. M., Monsan, P., & Boures, E. (1998). Enzyme and Microbial Technology, 23, 83–90.
Ferrer, M., Golyshina, O. V., Plou, F. J., Timmis, K. N., & Golyshin, P. N. (2005). Biochemical Journal, 391(Pt 2), 269–276.
Sheu, D. C., Duan, K. J., & Lin, C. T. (1994). Biotechnology Techniques, 8, 515–520.
Chen, G. G., Li, W., Zhang, Y. K., Qin, Y. L., Wu, K. Y., & Liang, Z. Q. (2011). World Journal of Microbiology and Biotechnology, 27, 1519–1523.
Schuster, E., Dunn-Coleman, N., Frisvad, J. C., & Van Dijck, P. W. (2002). Applied Microbiology and Biotechnology, 59(4–5), 426–435.
de Bekker, C., Wiebenga, A., Aguilar, G., & Wosten, H. A. (2009). Journal of Microbiological Methods, 76, 305–306.
Xu, D., Pan, L., Zhao, H., Zhao, M., Sun, J., & Liu, D. (2011). Journal of Industrial Microbiology and Biotechnology, 38(9), 1255–1265.
Zhu, F. M., Du, B., Gao, H. S., Liu, C. J., & Li, J. (2010). Prikladnaia Biokhimiia i Mikrobiologiia, 46(6), 678–684.
Scott, T. A., & Melvin, E. H. (1953). Analytical Chemistry, 25(11), 1656–1661.
Hida, H., Yamada, T., & Yamada, Y. (2007). Applied Microbiology and Biotechnology, 73, 1387–1393.
Xu, B., Jin, Z. H., Jin, Q. C., Li, N., & Cen, P. L. (2009). Biotechnology and Bioprocess Engineering, 14, 175–179.
van der Kaaij, R. M., Yuan, X. L., Franken, A., Ram, A. F., Punt, P. J., van der Maarel, M. J., & Dijkhuizen, L. (2007). Eukaryotic Cell, 6, 1178–1188.
Zhang, Y. X., Perry, K., Vinci, V. A., Powell, K., Stemmer, W. P., & del Cardayre, S. B. (2002). Nature, 415, 644–646.
Dai, M. H., Ziesman, S., Ratcliffe, T., Gill, R. T., & Copley, S. D. (2005). Metabolic Engineering, 7, 45–52.
John, R. P., Gangadharan, D., & Nampoothiri, K. M. (2008). Bioresource Technology, 99, 8008–8015.
Pel, H. J., de Winde, J. H., Archer, D. B., et al. (2007). Nature Biotechnology, 25, 221–231.
Nie, G., Yang, X., Liu, H., Wang, L., Gong, G., Jin, W., & Zheng, Z. (2013). Annals of Microbiology, 63(1), 279–287.
Siddique Awan, M., Tabbasam, N., Ayub, N., Babar, M. E., Mehboobur, R., Rana, S., & Rajoka, M. I. (2011). Molecular Biology Reports, 38(2), 1367–1374.
Wang, Y. H., Li, Y., Pei, X. L., Yu, L., & Feng, Y. (2007). Journal of Biotechnology, 129, 510–515.
Gong, J., Zheng, H., Wu, Z., Chen, T., & Zhao, X. (2009). Biotechnology Advances, 27(6), 996–1005.
Luo, J. M., Li, J. S., Liu, D., Liu, F., Wang, Y. T., Song, X. R., & Wang, M. (2012). Journal of Agricultural and Food Chemistry, 60, 6026–6036.
Zhao, M., Dai, C. C., Guan, X. Y., & Tao, J. (2009). Enzyme and Microbial Technology, 45(6–7), 419–425.
Lin, Q., Xiao, H., Zhao, J., Li, L., Yu, F., Liu, X., & Cheng, X. (2011). International Journal of Food Science and Technology, 46(6), 1194–1200.
Yun, J. W., Lee, M. G., & Song, S. K. (1994). Biotechnology Letters, 16, 1145–1150.
Zhang, L., Jiang, Y. J., Jiang, Z. Y., Sun, X. H., Shi, J. F., Cheng, W., & Sun, Q. Y. (2009). Biochemical Engineering Journal, 46, 186–192.
Lee, H. S., Auh, J. H., Yoon, H. G., Kim, M. J., Park, J. H., Hong, S. S., Kang, M. H., Kim, T. J., Moon, T. W., Kim, J. W., & Park, K. H. (2002). Journal of Agricultural and Food Chemistry, 50, 2812–2817.
Chen, W. C., Hung, T. F., & Lee, S. L. (1997). Biotechnology Letters, 19(10), 949–951.
Pan, Y. C., & Lee, W. C. (2005). Biotechnology and Bioengineering, 89(7), 797–804.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (grant no. 20362001 and 21062001), National “863” Research Program of China (grant no. 2006AA10Z339), Guangxi Natural Science Foundation (grant no. 2013GXNSFBA019074), and Sciences and Technology Development Fund of Guangxi Academy of Agricultural Sciences (grant no. 2013JZ05).
Author information
Authors and Affiliations
Corresponding author
Additional information
Wei Li and Guiguang Chen contributed equally to this work.
Rights and permissions
About this article
Cite this article
Li, W., Chen, G., Gu, L. et al. Genome Shuffling of Aspergillus niger for Improving Transglycosylation Activity. Appl Biochem Biotechnol 172, 50–61 (2014). https://doi.org/10.1007/s12010-013-0421-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-013-0421-x