Abstract
Solid-state fermentation has been widely used for enzyme production. However, secreted enzymes often bind to the solid substrate preventing their detection and recovery. A series of screening studies was performed to examine the role of extraction buffer composition including NaCl, ethylene glycol, sodium acetate buffer, and Tween 80, on xylanase and cellulase recovery from switchgrass. Our results indicated that the selection of an extraction buffer is highly dependent on the nature and source of the enzyme being extracted. While a buffer containing 50 mM sodium acetate at pH 5 was found to have a positive effect on the recovery of commercial fungal-derived cellulase and xylanase amended to switchgrass, the same buffer had a significant negative effect on enzyme extraction from solid fermentation samples colonized by the bacterium Acidothermus cellulolyticus. Xylanase activity was more affected by components in the extraction buffers compared to cellulase. This study demonstrated that extraction followed by diafiltration is important for assessing enzyme recovery from solid fermentation samples. Reduction in activity due to compounds present in the switchgrass extracts is reversible when the compounds are removed via diafiltration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acidothermus cellulolyticus is a highly thermotolerant hot spring bacterium that has been investigated as a source of enzymes for biomass hydrolysis. Aerobic, thermophilic decomposition of cellulose and hemicellulose by A. cellulolyticus has been confirmed experimentally [1, 2]. These features of A. cellulolyticus combined with its wide range of secreted enzymes make this organism a promising candidate for industrial biotechnology [3]. We have recently demonstrated production of xylanase and cellulase from solid-state fermentation (SSF) of switchgrass colonized by A. cellulolyticus [4]. However, when growing microorganisms on solid substrates for enzyme production, secreted enzymes may be trapped or bound to the solid substrate. Furthermore, bioconversion of lignocellulosic feedstocks like switchgrass via enzymatic hydrolysis and fermentation is affected by the presence and release of inhibitory compounds in lignocellulose and its hydrolysates [5]. Phenolic compounds at sufficient concentrations have been found to be toxic to cellulolytic microorganisms and enzymes [6–9]. Activities of xylanase, carboxymethylcellulase, and β-glucosidase produced by B. succinogenes were inhibited by phenolic monomers, especially p-coumaric acid [8]. An efficient extraction method that recovers bound enzymes and addresses co-extracted enzyme inhibitors is imperative for accurately evaluating enzyme secretion and yield.
Many methods have been used to extract enzymes from solid substrates colonized by microorganisms [10–13]. These methods involve extraction buffers that address non-covalent binding of proteins to solids. Interrupting binding during extraction is particularly important for enzymes that contain carbohydrate binding modules. The binding of a cellulase to cellulose was significantly improved by increasing the ionic strength of the buffer, suggesting that interaction with cellulose includes protonated groups on the enzyme [14]. Gupta et al. found ethylene glycol to be an effective eluent to release xylanase activity in hydrophobic interaction chromatography [15]. Ethylene glycol was also found to enhance xylanase thermal stability [15, 16]. Several studies suggest that Tween 80 and other surfactants reduce non-specific binding of cellulases to lignocellulose substrates, thus increasing enzymatic activity [17–21]. Tween 80 may also have a protective effect on cellulases to prevent their inactivation during hydrolysis [17, 18].
Despite this previous research, very little work has been done to examine the effect of different extraction buffer components on overall enzyme recovery from solid fermentation samples. Furthermore, enzymes originating from different sources (i.e., fungi or bacteria) might respond differently to extraction buffers. Therefore, in this research, a series of screening studies was performed to test the effect of different buffer components on cellulase and xylanase extraction from switchgrass amended with commercial enzymes (produced by fungi) as well as from switchgrass colonized by A. cellulolyticus. Three chemicals, NaCl, ethylene glycol, and Tween 80 that had been previously used in enzyme extraction studies were examined [19, 22, 23]. NaCl was used to address binding associated with a protonated group, ethylene glycol was examined to interrupt hydrophobic interactions, and Tween 80 was used to address non-specific binding [19]. These chemicals were added to sodium acetate buffer to maintain the pH at 5–6. During enzyme extraction from switchgrass colonized by A. cellulolyticus, the effect of sodium acetate buffer alone was also examined.
Materials and Methods
Microplate Enzyme Assays
Enzyme activities were assayed in a 96-well plate format (microplate assay). Samples were analyzed for xylanase activity on birchwood xylan substrate with xylose standards using a modified method based on a combination of Bailey et al. [24] and Xiao et al. [25] in a 96-well plate. Cellulase (endoglucanase) activity was measured using a similar method [25] with carboxymethyl cellulose (CMC) substrate and glucose standards. Both methods used an assay buffer consisting of 50 mM sodium acetate at pH 5.
The modified method used a PCR thermal cycler instead of a water bath for high throughput and uniform heat transfer during incubation and color development. Dinitrosalicylic acid (DNS) was used to detect reducing sugars formed in the enzyme-substrate reaction. For the enzyme assay, 40 μl of 1% birchwood xylan substrate or 2% CMC was added to each well in a 96-well PCR plate followed by 40 μl sample. The plate was sealed with foil, incubated (5 min for xylanase, 30 min for cellulase) at 50 °C, and the reactions were stopped by adding 80 μl DNS solution; standards were then added to the plate and also treated with DNS. The assay was completed by resealing the plate and heating to 95 °C (105 °C lid temperature to minimize condensation) for 5 min for color development. The absorbance at 540 nm was measured and a standard curve was constructed to calculate reducing sugars formed for each reaction.
Buffer Preparation, Addition of Enzymes to Switchgrass, and Extraction Procedures
A full-factorial experimental design was used to test the effect of NaCl, ethylene glycol, and Tween 80 on recovery of enzyme activity from switchgrass solids amended with mixed enzymes. Eight buffers were prepared (Table 1). For each trial, the ‘−1’ setting indicates the absence of the compound from the buffer and the ‘+1’ setting indicates levels of 0.9 wt.% NaCl, 0.1 wt.% Tween 80, and 25 wt.% ethylene glycol. All solutions were made in 50 mM sodium acetate buffer, which was the enzyme assay buffer, to maintain the pH at 5–6; therefore, trial 5 was solely 50 mM sodium acetate buffer.
The feedstock was switchgrass variety Trailblazer obtained from Dr. Dan Putnam at UC Davis. The approximate composition (dry basis) of this variety is 32% cellulose, 26% hemicellulose, and 18% lignin [26]. The switchgrass was oven-dried, milled with a knife mill, and passed through a 2-mm screen. Processed material was stored in a sealed container and maintained at room temperature until experimentation. Switchgrass was washed to reduce the background from monosaccharides in later reducing sugar assays. Washed switchgrass was prepared from 15 g dry switchgrass in 250-ml Erlenmeyer flasks. Washes were done by adding 150 g water to each flask with incubation for 1 h at room temperature and 150 rpm. Solids were collected by filtering the flask contents using 0.22-μm vacuum filtration units. Solids were rinsed from the flasks with 75 g water and filtered, giving an overall extraction ratio of 15:1 (g H2O:g switchgrass). Washed switchgrass retained by the filtration unit was air-dried for 48 h prior to addition of enzyme.
Xylanase (Dyadic Xylanase Plus) and cellulase (Novozymes Celluclast 1.5) were diluted in water to obtain a combined activity level of 0.53 IU/ml (μmol glucose released/min/ml) for cellulase and 5.9 IU/ml (μmol xylose released/min/ml) for xylanase. As received xylanase and cellulase (endoglucanase) activities were 32,000 IU/ml and 8.9 IU/ml, respectively. The enzyme mixture was added to washed switchgrass to bring the moisture content of the dried solids to approximately 80% wet basis. Wetted solids were incubated at room temperature for 1 h to allow enzyme-substrate binding. For each extraction, 3 g wet solids were added to 50-ml centrifuge tubes followed by 27 g extraction buffer. Samples were extracted for 1 h at 150 rpm at ambient temperature. Following extraction, tubes were centrifuged at 10,000 ×g and 4 °C for 10 min. Supernatant was collected and filtered using Nalgene 0.22-μm vacuum filtration units with polyethersulfone membrane (Nalge Nunc International). Aliquots of filtered supernatant were collected for enzyme assays and stored at 4 °C. The remaining supernatant samples were used for diafiltration using 5-kDa molecular weight cut-off Vivaspin spin columns (Sartorius Stedim Biotech, Goettingen, Germany) at 10,000 g and 4 °C. A total of 4 ml of the supernatant was added to the Vivaspin spin columns and centrifuged. Centrifugation was stopped every hour and the equivalent volume of liquid permeated was replaced with sodium acetate buffer before restarting centrifugation. A total of 4 ml of sodium acetate buffer was added to the column to replace the volume permeated. The centrifugation was continued to concentrate the buffer-exchanged sample. Retentate samples in the columns were then examined in xylanase and cellulase assays.
Buffer Preparation for Extraction from Switchgrass Colonized by A. cellulolyticus
A full-factorial experimental design with two mid-points was used to test the effect of NaCl, ethylene glycol, sodium acetate concentration (at pH 5), and Tween 80 on enzyme extraction from SSF of switchgrass colonized with A. cellulolyticus. As shown in Table 2, there were 18 trials in total. For each trial the ‘−1’ setting indicates the absence of the compound from the buffer, the ‘+1’ setting indicates levels of 0.9 wt.% NaCl, 0.1 wt.% Tween 80, 25 wt.% ethylene glycol, and 50 mM sodium acetate, which are commonly used in enzyme extraction from SSF [27–31], and the ‘0’ setting indicates (midpoint) levels of 0.45 wt.% NaCl, 0.05 wt.% Tween 80, 12.5 wt.% ethylene glycol, and 25 mM sodium acetate. Mid-points were used instead of replication to track any variation due to experimental errors.
Extraction of Enzymes from Switchgrass Colonized by A. cellulolyticus
The effectiveness of the selected buffers (Table 2) on extracting enzymes secreted during SSF was tested using switchgrass samples colonized by A. cellulolyticus. Prior to inoculation, switchgrass was prepared from 10 g dry switchgrass autoclaved for 30 min in 250-ml Erlenmeyer flasks. Switchgrass leaching was shown previously to be essential for growth of A. cellulolyticus on solid material [32]. Leaching was done by adding 150 g sterile water to each flask with incubation for 1 h at 55 °C and 150 rpm. Solids were collected by aseptically filtering the flask contents with 0.22-μm vacuum filtration units. Solids were rinsed from the flasks with 50 g sterile water and filtered, giving an overall extraction ratio of 20:1 (g H2O:g switchgrass) [32].
A. cellulolyticus inoculum was prepared in medium consisting of the following compounds in 1 l: 1 g NH4Cl, 1 g KH2PO4, 0.1 g MgSO4, 0.2 g CaCl2·2H2O, 1 g yeast extract, and 5 g cellobiose, with the pH adjusted to 5.2. Cultures were maintained in an orbital shaker at 55 °C and 150 rpm. Cells were harvested in the late log phase at an OD590 of approximately 0.40 and inoculated onto washed switchgrass retained on the filtration units at a ratio of 2 ml inoculum to 10 g dry switchgrass. After inoculation, the solids were mixed and amended with growth medium lacking cellobiose to bring the moisture content to about 80% wet basis and also provide nutrition for A. cellulolyticus. The inoculated solids were then transferred into 250-ml bioreactors [33] and incubated at 55 °C. Reactors were aerated continuously with humidified air at 5–7 ml/min to avoid oxygen limitations and prevent drying.
Enzymes were extracted from solid fermentation samples collected after 6 days of incubation using extraction buffers listed in Table 2. The extraction, centrifugation, and buffer exchange followed methods described earlier. Xylanase and cellulase activities were measured and compared.
HPLC Analysis
Supernatants were filtered through 0.22-μm syringe filters and analyzed for phenolic compounds by HPLC [34]. A Shimadzu Prominence HPLC (Columbia, MD) equipped with reversed-phase C-18 column (Agilent Zorbax SB-C18, 250 × 4.6 mm, 5 μm) and photo diode array detector was used. Solvent A was 2% aqueous acetic acid (v/v), Solvent B was methanol, the flow rate was 1 ml/min, and oven temperature was 30 °C. The following gradient of A in B (v/v) was used: 0–5 min = 100% A; 5–10 min = gradient 100–90% A; 10–15 min = 90% A; 15–20 min = gradient 90–80% A; 20–25 min = 80% A; 25–40 min = gradient 80–40% A; 40–50 min = gradient 40–20% A. The following eight phenolic compounds could efficiently be separated by HPLC [35]: p-hydroxybenzoic acid, p-hydroxybenzaldehyde, vanillic acid, syringic acid, vanillin, trans-p-coumaric acid, trans-ferulic acid, and hydrocinnamic acid. Phenolic standard mixtures containing 10, 50, 150, and 300 μg/ml of each of the eight compounds were used to estimate concentrations in the extracts using Shimadzu LC solution software.
Data Analysis:
The effects of different buffer components on the overall activity yield were examined using JMP IN v.7 (SAS Institute, Cary, NC) and fit using the following equation:
where E{Y} is the expected value of the response variable, β 0 is the model intercept, X 1 is the coded level (−1, 0, +1) for Tween 80, X 2 is the coded level (−1, 0, +1) for NaCl, X 3 is the coded level (−1, 0, +1) for ethylene glycol, X 4 is the coded level for sodium acetate (−1, 0, +1) and β 1, β 2, β 3, β 4 are the parameter estimates for the main effects of Tween 80, NaCl, ethylene glycol and sodium acetate, respectively. The parameters β 12, β 13, β 14, β 23, β 24, β 34 represent the interactions between Tween 80 and NaCl, Tween 80 and ethylene glycol, Tween 80 and sodium acetate, NaCl and ethylene glycol, NaCl and sodium acetate, and ethylene glycol and sodium acetate, respectively.
Reduced models were determined for xylanase and cellulase activity yields. Reduced models were developed using a mixed stepwise regression platform in JMP IN. JMP IN was also used to calculate significance probabilities (termed p values) for each parameter estimate. JMP IN’s “combine” rule was used to group each parameter estimate (e.g., β 12) with its precedent estimates (e.g., β 1 and β 2) and to calculate the group’s significance probability for entry as a joint F test. The significance probability for a parameter to enter and leave the model was set to 0.25.
Results and Discussion
Xylanase and Cellulase Activity Yield from Switchgrass Amended with a Mixture of Commercial Enzymes
Xylanase yields in switchgrass extracts before and after diafiltration are provided in Table 1. Treatments were normalized to trial 5 (sodium acetate buffer). Before diafiltration, all treatments resulted in an apparent lower enzyme recovery compared to sodium acetate buffer except for trial 6 (presence of NaCl), which had slightly higher recovery compared to sodium acetate alone. Diafiltration improved the activity for all treatments containing ethylene glycol except treatment 8 which contained all components (Table 1). In contrast, diafiltration lowered the enzyme activity for the trials containing NaCl, except for trial 7 where the presence of ethylene glycol likely dominated over the effects of other compounds. When all compounds were present in the buffer, diafiltration slightly lowered the xylanase activity, possibly as a result of NaCl removal. Upon examination of normalized xylanase activity before diafiltration, significant negative effects were found for Tween 80 (p value = 0.0025) and ethylene glycol (p value = 0.01) on xylanase activity (Table 3) based on their negative parameter estimates, whereas NaCl had a significant positive effect on the activity (p value = 0.02). After diafiltration, none of the chemicals had a significant effect on xylanase extraction efficiency (Table 3).
Normalized cellulase (endoglucanase) activity recovered from amended switchgrass is presented in Table 1. Without diafiltration, all treatments lowered the cellulase activity except for trial 4, where ethylene glycol was added to sodium acetate buffer. In general, however, diafiltration improved the activity of the cellulase for all treatments significantly (p value = 0.038) indicating that the activity of the cellulase was more affected by the presence of chemicals in the extraction buffer compared to xylanase.
Stepwise regression of normalized data was performed to identify critical components in the extraction buffer. Without diafiltration, Tween 80 showed a significant negative effect (p value = 0.01) (Table 3) on recovered cellulase activity. Following diafiltration, none of the chemicals had a significant effect on enzyme extraction efficiency, indicating that sodium acetate buffer alone could be the candidate buffer for extracting commercial enzymes from switchgrass.
Chemical additives were expected to improve the recovery of commercial enzymes amended to switchgrass. Our results indicated that without diafiltration these chemicals have a negative effect on xylanase and cellulase activity suggesting the need for diafiltration. After diafiltration, none of the examined chemicals significantly increased enzyme recovery (extraction efficiency) suggesting that sodium acetate buffer alone was most effective. In previous studies, acetate buffer at about pH 5 was used for enzyme extraction [29, 36]; however, since extraction buffer optimization was not the goal for these studies, effectiveness of acetate buffer was not justified on enzyme recovery and extraction.
Effect of Different Buffer Components on Xylanase and Cellulase Extraction from Switchgrass Colonized by A. cellulolyticus
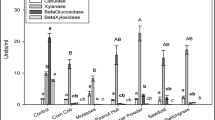
The effect of different extraction buffers on recovery of xylanase and cellulase produced during SSF of A. cellulolyticus was examined (Table 2). In general, xylanase activity varied with extraction buffer with or without diafiltration. Without diafiltration, all treatments containing sodium acetate had lower xylanase activity. NaCl had the same effect but not as profound as sodium acetate. NaCl (p value = 0.01) and sodium acetate (p value < 0.001) had significant negative effects on xylanase recovery, indicating an adverse effect of those compounds on xylanase activity (Table 4). Tween 80 had no significant effect on xylanase activity in extracts without diafiltration.
Although diafiltration significantly improved the measurement of xylanase activity extraction for all treatments, still NaCl (p value = 0.0042) and sodium acetate (0.0021) had the same negative effects on xylanase activity extraction. Acetate buffer was also found to be the worst extraction buffer for xylanase recovery elsewhere [27]. The negative effect of NaCl in our study suggested that the xylanases may have had binding modules to the surface of the plant polysaccharides which was adversely affected by the presence of ionic compounds. Improved xylanase activity after diafiltration could have been due to the removal of phenolic compounds and monosaccharides from the extract. Enzyme activity inhibition in the presence of phenolic compounds and monosaccharides has been investigated and shown previously [8, 37]. To confirm the effectiveness of diafiltration on removing phenolics and monosaccharides, four extracts were randomly selected from the 18 treatments and analyzed for the concentrations of phenolic compounds before and after diafiltration. As shown in Table 5, diafiltration removed the phenolic acids to levels below quantitation by the HPLC method. This result combined with higher enzyme activity after diafiltration confirmed the inhibitory effects of phenolic compounds (phenolic acids) and the reversibility of the enzyme inhibition after removing phenolic compounds. The average reducing sugar concentration in extracts was 0.5 ± 0.004 μmol/g switchgrass before diafiltration and it reduced to 0.14 ± 0.004 μmol/g switchgrass after diafiltration. While this concentration was lower after diafiltration, the concentration of monosaccharides in the extracts prior to diafiltration was likely too low to inhibit activity.
Due to inhibition, no cellulase activity was detected before diafiltration (data not shown). After diafiltration, all added chemicals had significant negative effects on cellulase activity except Tween 80 (Table 4). The results suggest that water is likely to be a satisfactory buffer for extracting enzymes in this particular situation. The effect of water on xylanase and cellulase extraction was not tested in enzyme amendment and extraction studies from switchgrass which concluded that sodium acetate buffer is the preferred extraction buffer. Comparison among the enzyme activities from switchgrass colonized by A. cellulolyticus extracted in the presence of 50 mM sodium acetate (treatments 5–7, 10, 12, 13, 16, and 17 in Table 2) indicated that 50 mM sodium acetate alone (treatment 17) has higher extraction efficiency compared to all other treatments that included 50 mM sodium acetate and other chemicals. This is in agreement with our finding in switchgrass amendment studies. Both sodium acetate buffer and water have been introduced as preferable buffers for xylanase extraction from SSF [29, 36, 38–40]; however, inhibition of enzymes by buffer components was not examined. It should be mentioned that fungal enzymes were used in those studies in contrast with present study where bacterial enzyme was produced, therefore, it is likely to observe some differences in the nature of the enzyme and consequently in the extraction optimization.
Other researchers have studied the effects of Tween 80 and NaCl on extraction of enzymes from SSF. Diaz et al. [31] examined the effect of Tween 80, NaCl, and other components on extraction of xylanase from fermented grape pomace. Aqueous Tween 80 gave the highest xylanase activity, a 66% improvement over extraction with water; the authors attribute this to an increase in cell permeability, increasing the export of molecules across the cell membrane. They found NaCl gave a 35% improvement compared to water, but did not recommend NaCl for xylanase extraction or test any combinations. Other researchers found that Tween 80 in physiological saline was no better than sodium acetate buffer in extracting xylanase from sugarcane bagasse, but repetitive extractions improved overall activity yield [30]. Although our results with Tween 80 were somewhat similar to the reported studies, our finding with NaCl contradicted reported results, indicating again that specific enzymes, microorganisms, and feedstocks are likely to affect the overall extraction efficiency and effectiveness. Furthermore, typical reaction conditions are held constant when measuring the activity of an enzyme on a given substrate. Changes in pH and temperature can alter the specific activity of an enzyme. Likewise, changes in ionic strength or hydrophobicity can potentially affect binding of the enzymes to the substrate. These and other factors could affect the enzyme assays.
Commercial enzymes responded differently to extraction buffers as compared to the enzymes secreted by A. cellulolyticus. This was most probably due to the differences in their nature since the effectiveness of an extraction buffer on enzyme recovery depends on type of enzyme, source of enzyme and substrate being used [27]. Commercial enzymes used in this study had fungal origin, whereas the enzymes recovered from SSF samples had bacterial origin. Furthermore, commercial enzymes have been through product recovery and stabilization steps that might have changed their chemical properties and interactions with other chemicals. More studies are needed to prove this hypothesis.
Conclusions
In this study, the effectiveness of four chemicals as extraction buffers was tested on enzyme activity and recovery from switchgrass. The results from this study showed that the nature and source of the enzyme likely play an important role on extracting enzymes from solid substrates. For switchgrass samples colonized by A. cellulolyticus, water was found to be the preferred extraction buffer, whereas for commercial fungal-derived enzymes amended to switchgrass 50 mM sodium acetate was the ideal buffer. Although none of the added chemicals showed a significant positive effect on enzyme extraction, diafiltration was an imperative step to recover more enzyme activity. Results suggest that diafiltration improved the activity by removing inhibitory phenolic compounds from the extract. In order to achieve more concentrated crude enzyme from switchgrass colonized by A. cellulolyticus, further studies are needed to examine the effects of temperature, agitation, and extraction time.
References
Maréchal, J., Clement, B., Nalin, R., Gandon, R., Orso, S., Cvejic, H., et al. (2000). International Journal of Systematic and Evolutionary Microbiology, 50, 781–785.
Mohagheghi, A., Grohmann, K. M. M. H., Leighton, L., & Updegraff, D. M. (1986). J. Syst. Bacteriol., 36, 435–443.
Sakon, J., Adney, W. S., Himmel, M. E., Thomas, S. R., & Karplus, P. A. (1996). Biochemistry, 35, 10648–10660.
Rezaei, F., Joh, L., Berry, A., & VanderGheynst, J. S. (2011). Biofuels, 2(1), 21–32.
Palmqvist, E., & Hahn-Hagerdal, B. (2000). Bioresource Technology, 74, 17–24.
Borneman, W. S., Akin, D. E., & Vaneseltine, W. P. (1986). Applied and Environmental Microbiology, 52, 1331–1339.
Chesson, A., Stewart, C. S., & Wallace, R. J. (1982). Applied and Environmental Microbiology, 44, 597–603.
Martin, S. A., & Akin, D. E. (1988). Applied and Environmental Microbiology, 54, 3019–3022.
Scalbert, A. (1991). Phytochemistry, 30, 3875–3883.
Archana, A., & Satyanarayana, T. (1997). Enzyme and Microbial Technology, 21, 12–17.
Castilho, L. R., Medronho, R. A., & Alves, T. L. M. (2000). Bioresource Technology, 71, 45–50.
Khandeparkar, R. D. S., & Bhosle, N. B. (2006). Enzyme and Microbial Technology, 39, 732–742.
Krishna, C. (1999). Bioresource Technology, 69, 231–239.
Reinikainen, T., Teleman, O., & Teeri, T. T. (1995). Proteins, 22, 392–403.
Gupta, A., Roy, I., Khare, S. K., Bisaria, V. S., & Gupta, M. N. (2002). Biotechnological Letters, 24, 2005–2009.
Cobos, A., & Estrada, P. (2003). Enzyme and Microbial Technology, 33, 810–818.
Eriksson, T., Borjesson, J., & Tjerneld, F. (2002). Enzyme and Microbial Technology, 31, 353–364.
Helle, S. S., Duff, S. J. B., & Cooper, D. G. (1993). Biotechnology and Bioengineering, 42, 611–617.
Otter, D. E., Munro, P. A., Scott, G. K., & Geddes, R. (1989). Biotechnology and Bioengineering, 34, 291–298.
Xu, F., Ding, H. S., Osborn, D., Tejirian, A., Brown, K., Albano, W., et al. (2008). Journal of Molecular Catalysis. B, Enzymatic, 51, 42–48.
Zheng, Y., Pan, Z. L., Zhang, R. H., Wang, D. H., & Jenkins, B. (2008). Applied Biochemistry and Biotechnology, 146, 231–248.
Szakács, G., Urbánszki, K., & Tengerdy Robert, P. (2000). Glycosyl hydrolases for biomass conversion, vol solid-state enzymes for fiber hydrolysis (pp. 190–203). Washington DC: American Chemical Society.
Maciel, G. M., de-Souza-Vandenberghe, L. P., Isidoro-Haminiuk, C. W., Fendrich, R. C., Della-Bianca, B. E., da-Silva-Brandalize, T. Q., et al. (2008). Food Technology and Biotechnology, 46, 183–189.
Bailey, M. J., Biely, P., & Poutanen, K. (1992). Journal of Biotechnology, 23, 257–270.
Xiao, Z. Z., Storms, R., & Tsang, A. (2005). Analytical Biochemistry, 342, 176–178.
Keshwani, D. R., & Cheng, J. J. (2009). Bioresource Technology, 100, 1515–1523.
Maciel, G., de Souza Vandenberghe, L., Fendrich, R., Della Bianca, B., Haminiuk, C., & Soccol, C. (2009). Biotechnology and Bioprocess Engineering, 14, 748–755.
Pal, A., & Khanum, F. (2010). Bioresource Technology, 101, 7563–7569.
de Souza, M. C., Roberto, I. C., & Milagres, A. M. F. (1999). Applied Microbiology and Biotechnology, 52, 768–772.
Rezende, M. I., de Barbosa, A. M., Vasconcelos, A. F. D., & Endo, A. S. (2002). Braz J Microbiol, 33, 67–72.
Diaz, A. B., Caro, I., de Ory, I., & Blandino, A. (2007). Enzyme and Microbial Technology, 41, 302–306.
VanderGheynst, J. S., Rezaei, F., Dooley, T. M., & Berry, A. M. (2010). Biotechnology Progress, 26, 622–626.
May, B. A., & VanderGheynst, J. S. (2001). Journal of Industrial Microbiology & Biotechnology, 27, 203–207.
Arimboor, R., Kumar, K. S., & Arumughan, C. (2008). Journal of Pharmaceutical and Biomedical Analysis, 47, 31–38.
Joh, L. D., Rezaei, F., Parales, J. V., Parales, R. E., Barabote, R. D., Berry, A. M., et al. (2010). Biotechnol Prog. doi:10.1002/btpr.525.
Silva, C. H. C., Puls, J., de Sousa, M. V., & Ferreira Filho, E. X. (1999). Revista de Microbiologia, 30, 114–119.
Philippidis, G. P., Smith, T. K., & Wyman, C. E. (1993). Biotechnology and Bioengineering, 41, 846–853.
Liu, W., Zhu, W., Lu, Y., Kong, J., & Ma, G. (1998). Process Biochemistry, 33, 331–336.
Heck, J. X., Hertz, P. F., & Ayub, M. A. Z. (2002). Braz J Microbiol, 33, 213–218.
Heck, J. X., Hertz, P. F., & Ayub, M. A. Z. (2005). Process Biochemistry, 40, 2891–2895.
Acknowledgments
Funding for this research was provided by Chevron Technology Ventures. The authors wish to thank Christopher Lee and Joshua Claypool for assistance with enzyme extraction studies. Research by A.P. Reddy and J.S. VanderGheynst was performed as part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Rezaei, F., Joh, L.D., Kashima, H. et al. Selection of Conditions for Cellulase and Xylanase Extraction from Switchgrass Colonized by Acidothermus cellulolyticus . Appl Biochem Biotechnol 164, 793–803 (2011). https://doi.org/10.1007/s12010-011-9174-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-011-9174-6