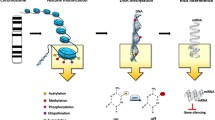
Abstract
Purpose of Review
Epigenetics has been implicated in the pathogenesis of systemic sclerosis (SSc). In this review, the involvement of the three epigenetic mechanisms in SSc development and progression—DNA methylation, histone modifications, and non-coding RNAs—will be discussed.
Recent Findings
Alteration in epigenetics was observed in immune cells, dermal fibroblasts, and endothelial cells derived from SSc patients. Genes that are affected include those involved in immune cell function and differentiation, TGFβ and Wnt pathways, extracellular matrix accumulation, transcription factors, and angiogenesis. All the studies remain in the pre-clinical stage.
Summary
Extensive research provides evidence that epigenetic alterations are critical for SSc pathogenesis. Future epigenomic studies will undoubtedly continue to broaden our understanding of disease pathogenesis and clinical heterogeneity. They will also provide the scientific basis for repurposing epigenetic-modifying agents for SSc patients.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Tsou PS, Sawalha AH. Unfolding the pathogenesis of scleroderma through genomics and epigenomics. J Autoimmun. 2017;83:73–94. https://doi.org/10.1016/j.jaut.2017.05.004.
Salazar G, Mayes MD. Genetics, epigenetics, and genomics of systemic sclerosis. Rheum Dis Clin N Am. 2015;41(3):345–66. https://doi.org/10.1016/j.rdc.2015.04.001.
Arnett FC, Cho M, Chatterjee S, Aguilar MB, Reveille JD, Mayes MD. Familial occurrence frequencies and relative risks for systemic sclerosis (scleroderma) in three United States cohorts. Arthritis Rheum. 2001;44(6):1359–62. https://doi.org/10.1002/1529-0131(200106)44:6<1359::AID-ART228>3.0.CO;2-S.
Feghali-Bostwick C, Medsger TA Jr, Wright TM. Analysis of systemic sclerosis in twins reveals low concordance for disease and high concordance for the presence of antinuclear antibodies. Arthritis Rheum. 2003;48(7):1956–63. https://doi.org/10.1002/art.11173.
McCormic ZD, Khuder SS, Aryal BK, Ames AL, Khuder SA. Occupational silica exposure as a risk factor for scleroderma: a meta-analysis. Int Arch Occup Environ Health. 2010;83(7):763–9. https://doi.org/10.1007/s00420-009-0505-7.
De Decker E, Vanthuyne M, Blockmans D, Houssiau F, Lenaerts J, Westhovens R, et al. High prevalence of occupational exposure to solvents or silica in male systemic sclerosis patients: a Belgian cohort analysis. Clin Rheumatol. 2018;37(7):1977–82. https://doi.org/10.1007/s10067-018-4045-y.
Marie I, Gehanno JF, Bubenheim M, Duval-Modeste AB, Joly P, Dominique S, et al. Systemic sclerosis and exposure to heavy metals: a case control study of 100 patients and 300 controls. Autoimmun Rev. 2017;16(3):223–30. https://doi.org/10.1016/j.autrev.2017.01.004.
Moroncini G, Mori S, Tonnini C, Gabrielli A. Role of viral infections in the etiopathogenesis of systemic sclerosis. Clin Exp Rheumatol. 2013;31(2 Suppl 76):3–7.
Haustein UF, Haupt B. Drug-induced scleroderma and sclerodermiform conditions. Clin Dermatol. 1998;16(3):353–66.
Rezaei R, Mahmoudi M, Gharibdoost F, Kavosi H, Dashti N, Imeni V, et al. IRF7 gene expression profile and methylation of its promoter region in patients with systemic sclerosis. Int J Rheum Dis. 2017;20(10):1551–61. https://doi.org/10.1111/1756-185X.13175.
• Zhu H, Zhu C, Mi W, Chen T, Zhao H, Zuo X, et al. Integration of genome-wide DNA methylation and transcription uncovered aberrant methylation-regulated genes and pathways in the peripheral blood mononuclear cells of systemic sclerosis. Int J Rheumatol. 2018;2018:7342472. https://doi.org/10.1155/2018/7342472. Identified methylation-regulated genes in scleroderma PBMCs.
Chu H, Jiang S, Liu Q, Ma Y, Zhu X, Liang M, et al. Sirtuin1 protects against systemic sclerosis-related pulmonary fibrosis by decreasing proinflammatory and profibrotic processes. Am J Respir Cell Mol Biol. 2018;58(1):28–39. https://doi.org/10.1165/rcmb.2016-0192OC.
Dolcino M, Tinazzi E, Puccetti A, Lunardi C. In systemic sclerosis, a unique long non coding RNA regulates genes and pathways involved in the three main features of the disease (vasculopathy, fibrosis and autoimmunity) and in carcinogenesis. J Clin Med. 2019;8(3). https://doi.org/10.3390/jcm8030320.
Lei W, Luo Y, Lei W, Luo Y, Yan K, Zhao S, et al. Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand J Rheumatol. 2009;38(5):369–74. https://doi.org/10.1080/03009740902758875.
Jiang H, Xiao R, Lian X, Kanekura T, Luo Y, Yin Y, et al. Demethylation of TNFSF7 contributes to CD70 overexpression in CD4+ T cells from patients with systemic sclerosis. Clin Immunol. 2012;143(1):39–44. https://doi.org/10.1016/j.clim.2012.01.005.
Lian X, Xiao R, Hu X, Kanekura T, Jiang H, Li Y, et al. DNA demethylation of CD40l in CD4+ T cells from women with systemic sclerosis: a possible explanation for female susceptibility. Arthritis Rheum. 2012;64(7):2338–45. https://doi.org/10.1002/art.34376.
Wang Y, Shu Y, Xiao Y, Wang Q, Kanekura T, Li Y, et al. Hypomethylation and overexpression of ITGAL (CD11a) in CD4(+) T cells in systemic sclerosis. Clin Epigenetics. 2014;6(1):25. https://doi.org/10.1186/1868-7083-6-25.
•• Ding W, Pu W, Wang L, Jiang S, Zhou X, Tu W, et al. Genome-wide DNA methylation analysis in systemic sclerosis reveals hypomethylation of IFN-associated genes in CD4(+) and CD8(+) T cells. J Invest Dermatol. 2018;138(5):1069–77. https://doi.org/10.1016/j.jid.2017.12.003. Describes genome-wide DNA methylation chages in CD4+ and CD8+ T cells from scleroderma patients.
Wang YY, Wang Q, Sun XH, Liu RZ, Shu Y, Kanekura T, et al. DNA hypermethylation of the forkhead box protein 3 (FOXP3) promoter in CD4+ T cells of patients with systemic sclerosis. Br J Dermatol. 2014;171(1):39–47. https://doi.org/10.1111/bjd.12913.
Ugor E, Simon D, Almanzar G, Pap R, Najbauer J, Nemeth P, et al. Increased proportions of functionally impaired regulatory T cell subsets in systemic sclerosis. Clin Immunol. 2017;184:54–62. https://doi.org/10.1016/j.clim.2017.05.013.
Wang Q, Xiao Y, Shi Y, Luo Y, Li Y, Zhao M, et al. Overexpression of JMJD3 may contribute to demethylation of H3K27me3 in CD4+ T cells from patients with systemic sclerosis. Clin Immunol. 2015;161(2):396–9. https://doi.org/10.1016/j.clim.2015.03.006.
Wang Y, Yang Y, Luo Y, Yin Y, Wang Q, Li Y, et al. Aberrant histone modification in peripheral blood B cells from patients with systemic sclerosis. Clin Immunol. 2013;149(1):46–54. https://doi.org/10.1016/j.clim.2013.06.006.
•• van der Kroef M, Castellucci M, Mokry M, Cossu M, Garonzi M, Bossini-Castillo LM, et al. Histone modifications underlie monocyte dysregulation in patients with systemic sclerosis, underlining the treatment potential of epigenetic targeting. Ann Rheum Dis. 2019;78(4):529–38. https://doi.org/10.1136/annrheumdis-2018-214295. First study examining histone changes in monocytes from scleroderma patients.
Mariotti B, Servaas NH, Rossato M, Tamassia N, Cassatella MA, Cossu M, et al. The long non-coding RNA NRIR drives IFN-response in monocytes: implication for systemic sclerosis. Front Immunol. 2019;10:100. https://doi.org/10.3389/fimmu.2019.00100.
Ciechomska M, Zarecki P, Merdas M, Swierkot J, Morgiel E, Wiland P, et al. The role of microRNA-5196 in the pathogenesis of systemic sclerosis. Eur J Clin Investig. 2017;47(8):555–64. https://doi.org/10.1111/eci.12776.
• Rossato M, Affandi AJ, Thordardottir S, Wichers CGK, Cossu M, Broen JCA, et al. Association of microRNA-618 expression with altered frequency and activation of plasmacytoid dendritic cells in patients with systemic sclerosis. Arthritis Rheum. 2017;69(9):1891–902. https://doi.org/10.1002/art.40163. First study examining epigenetic changes in plasmacytoid dendritic cells in scleroderma.
Hattori M, Yokoyama Y, Hattori T, Motegi S, Amano H, Hatada I, et al. Global DNA hypomethylation and hypoxia-induced expression of the ten eleven translocation (TET) family, TET1, in scleroderma fibroblasts. Exp Dermatol. 2015;24(11):841–6. https://doi.org/10.1111/exd.12767.
• Altorok N, Tsou PS, Coit P, Khanna D, Sawalha AH. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann Rheum Dis. 2015;74(8):1612–20. https://doi.org/10.1136/annrheumdis-2014-205303. First study to utilize DNA methylation arrays to examine the methylome in dermal fibroblasts isolated from healthy controls and scleroderma patients.
Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. 2006;54(7):2271–9. https://doi.org/10.1002/art.21948.
Noda S, Asano Y, Nishimura S, Taniguchi T, Fujiu K, Manabe I, et al. Simultaneous downregulation of KLF5 and Fli1 is a key feature underlying systemic sclerosis. Nat Commun. 2014;5:5797. https://doi.org/10.1038/ncomms6797.
Zhang Y, Potter S, Chen CW, Liang R, Gelse K, Ludolph I, et al. Poly(ADP-ribose) polymerase-1 regulates fibroblast activation in systemic sclerosis. Ann Rheum Dis. 2018;77(5):744–51. https://doi.org/10.1136/annrheumdis-2017-212265.
Henderson J, Brown M, Horsburgh S, Duffy L, Wilkinson S, Worrell J, et al. Methyl cap binding protein 2: a key epigenetic protein in systemic sclerosis. Rheumatology (Oxford). 2018. https://doi.org/10.1093/rheumatology/key327.
• He Y, Tsou PS, Khanna D, Sawalha AH. Methyl-CpG-binding protein 2 mediates antifibrotic effects in scleroderma fibroblasts. Ann Rheum Dis. 2018;77(8):1208–18. https://doi.org/10.1136/annrheumdis-2018-213022. Mechanistic study to determine the role of MeCP2 in scleroderma fibrosis utilizing RNA-seq and ChIP-seq.
Kramer M, Dees C, Huang J, Schlottmann I, Palumbo-Zerr K, Zerr P, et al. Inhibition of H3K27 histone trimethylation activates fibroblasts and induces fibrosis. Ann Rheum Dis. 2013;72(4):614–20. https://doi.org/10.1136/annrheumdis-2012-201615.
•• Tsou PS, Campbell P, Amin MA, Coit P, Miller S, Fox DA, et al. Inhibition of EZH2 prevents fibrosis and restores normal angiogenesis in scleroderma. Proc Natl Acad Sci U S A. 2019;116(9):3695–702. https://doi.org/10.1073/pnas.1813006116. First study suggesting the impact of histone methylation in scleroderma endothelial cells.
Bergmann C, Brandt A, Merlevede B, Hallenberger L, Dees C, Wohlfahrt T, et al. The histone demethylase Jumonji domain-containing protein 3 (JMJD3) regulates fibroblast activation in systemic sclerosis. Ann Rheum Dis. 2018;77(1):150–8. https://doi.org/10.1136/annrheumdis-2017-211501.
Ghosh AK, Bhattacharyya S, Lafyatis R, Farina G, Yu J, Thimmapaya B, et al. p300 is elevated in systemic sclerosis and its expression is positively regulated by TGF-beta: epigenetic feed-forward amplification of fibrosis. J Invest Dermatol. 2013;133(5):1302–10. https://doi.org/10.1038/jid.2012.479.
•• Shin JY, Beckett JD, Bagirzadeh R, Creamer TJ, Shah AA, McMahan Z et al. Epigenetic activation and memory at a TGFB2 enhancer in systemic sclerosis. Sci Transl Med. 2019;11(497). https://doi.org/10.1126/scitranslmed.aaw0790. Comprehensive examination of epigenetic control of TGFB2 enhancer in SSc dermal fibroblasts utlizing RNA-seq, ATAC-seq, and genome-editing techniques.
Wei J, Ghosh AK, Chu H, Fang F, Hinchcliff ME, Wang J, et al. The histone deacetylase sirtuin 1 is reduced in systemic sclerosis and abrogates fibrotic responses by targeting transforming growth factor beta signaling. Arthritis Rheum. 2015;67(5):1323–34. https://doi.org/10.1002/art.39061.
Zerr P, Palumbo-Zerr K, Huang J, Tomcik M, Sumova B, Distler O, et al. Sirt1 regulates canonical TGF-β signalling to control fibroblast activation and tissue fibrosis. Ann Rheum Dis. 2014. https://doi.org/10.1136/annrheumdis-2014-205740.
Akamata K, Wei J, Bhattacharyya M, Cheresh P, Bonner MY, Arbiser JL, et al. SIRT3 is attenuated in systemic sclerosis skin and lungs, and its pharmacologic activation mitigates organ fibrosis. Oncotarget. 2016;7(43):69321–36. https://doi.org/10.18632/oncotarget.12504.
Maurer B, Stanczyk J, Jungel A, Akhmetshina A, Trenkmann M, Brock M, et al. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum. 2010;62(6):1733–43. https://doi.org/10.1002/art.27443.
Honda N, Jinnin M, Kajihara I, Makino T, Makino K, Masuguchi S, et al. TGF-beta-mediated downregulation of microRNA-196a contributes to the constitutive upregulated type I collagen expression in scleroderma dermal fibroblasts. J Immunol. 2012;188(7):3323–31. https://doi.org/10.4049/jimmunol.1100876.
Makino K, Jinnin M, Hirano A, Yamane K, Eto M, Kusano T, et al. The downregulation of microRNA let-7a contributes to the excessive expression of type I collagen in systemic and localized scleroderma. J Immunol. 2013;190(8):3905–15. https://doi.org/10.4049/jimmunol.1200822.
Jafarinejad-Farsangi S, Farazmand A, Mahmoudi M, Gharibdoost F, Karimizadeh E, Noorbakhsh F, et al. MicroRNA-29a induces apoptosis via increasing the Bax:Bcl-2 ratio in dermal fibroblasts of patients with systemic sclerosis. Autoimmunity. 2015;48(6):369–78. https://doi.org/10.3109/08916934.2015.1030616.
Ciechomska M, O'Reilly S, Suwara M, Bogunia-Kubik K, van Laar JM. MiR-29a reduces TIMP-1 production by dermal fibroblasts via targeting TGF-beta activated kinase 1 binding protein 1, implications for systemic sclerosis. PLoS One. 2014;9(12):e115596. https://doi.org/10.1371/journal.pone.0115596.
Honda N, Jinnin M, Kira-Etoh T, Makino K, Kajihara I, Makino T, et al. miR-150 down-regulation contributes to the constitutive type I collagen overexpression in scleroderma dermal fibroblasts via the induction of integrin beta3. Am J Pathol. 2013;182(1):206–16. https://doi.org/10.1016/j.ajpath.2012.09.023.
Zhu H, Luo H, Li Y, Zhou Y, Jiang Y, Chai J, et al. MicroRNA-21 in scleroderma fibrosis and its function in TGF-beta-regulated fibrosis-related genes expression. J Clin Immunol. 2013;33(6):1100–9. https://doi.org/10.1007/s10875-013-9896-z.
Zhu H, Li Y, Qu S, Luo H, Zhou Y, Wang Y, et al. MicroRNA expression abnormalities in limited cutaneous scleroderma and diffuse cutaneous scleroderma. J Clin Immunol. 2012;32(3):514–22. https://doi.org/10.1007/s10875-011-9647-y.
Yan Q, Chen J, Li W, Bao C, Fu Q. Targeting miR-155 to treat experimental scleroderma. Sci Rep. 2016;6:20314. https://doi.org/10.1038/srep20314.
Artlett CM, Sassi-Gaha S, Hope JL, Feghali-Bostwick CA, Katsikis PD. Mir-155 is overexpressed in systemic sclerosis fibroblasts and is required for NLRP3 inflammasome-mediated collagen synthesis during fibrosis. Arthritis Res Ther. 2017;19(1):144. https://doi.org/10.1186/s13075-017-1331-z.
Zhou B, Zhu H, Luo H, Gao S, Dai X, Li Y, et al. MicroRNA-202-3p regulates scleroderma fibrosis by targeting matrix metalloproteinase 1. Biomed Pharmacother. 2017;87:412–8. https://doi.org/10.1016/j.biopha.2016.12.080.
Sing T, Jinnin M, Yamane K, Honda N, Makino K, Kajihara I, et al. microRNA-92a expression in the sera and dermal fibroblasts increases in patients with scleroderma. Rheumatology (Oxford). 2012;51(9):1550–6. https://doi.org/10.1093/rheumatology/kes120.
O'Reilly S, Ciechomska M, Fullard N, Przyborski S, van Laar JM. IL-13 mediates collagen deposition via STAT6 and microRNA-135b: a role for epigenetics. Sci Rep. 2016;6:25066. https://doi.org/10.1038/srep25066.
Luo H, Zhu H, Zhou B, Xiao X, Zuo X. MicroRNA-130b regulates scleroderma fibrosis by targeting peroxisome proliferator-activated receptor gamma. Mod Rheumatol. 2015;25(4):595–602. https://doi.org/10.3109/14397595.2014.1001311.
Liakouli V, Cipriani P, Di Benedetto P, Panzera N, Ruscitti P, Pantano I, et al. Epidermal growth factor like-domain 7 and miR-126 are abnormally expressed in diffuse systemic sclerosis fibroblasts. Sci Rep. 2019;9(1):4589. https://doi.org/10.1038/s41598-019-39485-8.
Alsaleh G, Francois A, Philippe L, Gong YZ, Bahram S, Cetin S, et al. MiR-30a-3p negatively regulates BAFF synthesis in systemic sclerosis and rheumatoid arthritis fibroblasts. PLoS One. 2014;9(10):e111266. https://doi.org/10.1371/journal.pone.0111266.
Iwamoto N, Vettori S, Maurer B, Brock M, Pachera E, Jungel A, et al. Downregulation of miR-193b in systemic sclerosis regulates the proliferative vasculopathy by urokinase-type plasminogen activator expression. Ann Rheum Dis. 2016;75(1):303–10. https://doi.org/10.1136/annrheumdis-2014-205326.
Nakamura K, Jinnin M, Harada M, Kudo H, Nakayama W, Inoue K, et al. Altered expression of CD63 and exosomes in scleroderma dermal fibroblasts. J Dermatol Sci. 2016;84(1):30–9. https://doi.org/10.1016/j.jdermsci.2016.06.013.
Wang Z, Jinnin M, Nakamura K, Harada M, Kudo H, Nakayama W, et al. Long non-coding RNA TSIX is upregulated in scleroderma dermal fibroblasts and controls collagen mRNA stabilization. Exp Dermatol. 2016;25(2):131–6. https://doi.org/10.1111/exd.12900.
• Messemaker TC, Chadli L, Cai G, Goelela VS, Boonstra M, Dorjee AL, et al. Antisense long non-coding RNAs are deregulated in skin tissue of patients with systemic sclerosis. J Invest Dermatol. 2018;138(4):826–35. https://doi.org/10.1016/j.jid.2017.09.053. Identification of three novel long non-coding RNAs in scleroderma skin.
Wyman AE, Noor Z, Fishelevich R, Lockatell V, Shah NG, Todd NW, et al. Sirtuin 7 is decreased in pulmonary fibrosis and regulates the fibrotic phenotype of lung fibroblasts. Am J Phys Lung Cell Mol Phys. 2017;312(6):L945–l58. https://doi.org/10.1152/ajplung.00473.2016.
Wang Y, Kahaleh B. Epigenetic repression of bone morphogenetic protein receptor II expression in scleroderma. J Cell Mol Med. 2013. https://doi.org/10.1111/jcmm.12105.
•• Tsou PS, Wren JD, Amin MA, Schiopu E, Fox DA, Khanna D, et al. Histone deacetylase 5 is overexpressed in scleroderma endothelial cells and impairs angiogenesis via repression of proangiogenic factors. Arthritis Rheum. 2016;68(12):2975–85. https://doi.org/10.1002/art.39828. Utilizing ATAC-seq to identify HDAC5-target genes in scleroderma ECs.
Dashti N, Mahmoudi M, Gharibdoost F, Kavosi H, Rezaei R, Imeni V, et al. Evaluation of ITGB2 (CD18) and SELL (CD62L) genes expression and methylation of ITGB2 promoter region in patients with systemic sclerosis. Rheumatol Int. 2018;38(3):489–98. https://doi.org/10.1007/s00296-017-3915-y.
Dees C, Schlottmann I, Funke R, Distler A, Palumbo-Zerr K, Zerr P, et al. The Wnt antagonists DKK1 and SFRP1 are downregulated by promoter hypermethylation in systemic sclerosis. Ann Rheum Dis. 2013. https://doi.org/10.1136/annrheumdis-2012-203194.
Svegliati S, Marrone G, Pezone A, Spadoni T, Grieco A, Moroncini G, et al. Oxidative DNA damage induces the ATM-mediated transcriptional suppression of the Wnt inhibitor WIF-1 in systemic sclerosis and fibrosis. Sci Signal. 2014;7(341):ra84. https://doi.org/10.1126/scisignal.2004592.
Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25(11):4727–41. https://doi.org/10.1128/MCB.25.11.4727-4741.2005.
Qi Q, Guo Q, Tan G, Mao Y, Tang H, Zhou C, et al. Predictors of the scleroderma phenotype in fibroblasts from systemic sclerosis patients. J Eur Acad Dermatol Venereol: JEADV. 2009;23(2):160–8. https://doi.org/10.1111/j.1468-3083.2008.03016.x.
Kubo M, Czuwara-Ladykowska J, Moussa O, Markiewicz M, Smith E, Silver RM, et al. Persistent down-regulation of Fli1, a suppressor of collagen transcription, in fibrotic scleroderma skin. Am J Pathol. 2003;163(2):571–81. https://doi.org/10.1016/s0002-9440(10)63685-1.
Asano Y. Epigenetic suppression of Fli1, a potential predisposing factor in the pathogenesis of systemic sclerosis. Int J Biochem Cell Biol. 2015;67:86–91. https://doi.org/10.1016/j.biocel.2015.06.004.
Yan J, Li B, Lin B, Lee PT, Chung TH, Tan J, et al. EZH2 phosphorylation by JAK3 mediates a switch to noncanonical function in natural killer/T-cell lymphoma. Blood. 2016;128(7):948–58. https://doi.org/10.1182/blood-2016-01-690701.
Ghosh AK, Varga J. The transcriptional coactivator and acetyltransferase p300 in fibroblast biology and fibrosis. J Cell Physiol. 2007;213(3):663–71. https://doi.org/10.1002/jcp.21162.
Bhattacharyya S, Ghosh AK, Pannu J, Mori Y, Takagawa S, Chen G, et al. Fibroblast expression of the coactivator p300 governs the intensity of profibrotic response to transforming growth factor beta. Arthritis Rheum. 2005;52(4):1248–58. https://doi.org/10.1002/art.20996.
Huber LC, Distler JH, Moritz F, Hemmatazad H, Hauser T, Michel BA, et al. Trichostatin A prevents the accumulation of extracellular matrix in a mouse model of bleomycin-induced skin fibrosis. Arthritis Rheum. 2007;56(8):2755–64. https://doi.org/10.1002/art.22759.
Hemmatazad H, Rodrigues HM, Maurer B, Brentano F, Pileckyte M, Distler JH, et al. Histone deacetylase 7, a potential target for the antifibrotic treatment of systemic sclerosis. Arthritis Rheum. 2009;60(5):1519–29. https://doi.org/10.1002/art.24494.
Sosulski ML, Gongora R, Feghali-Bostwick C, Lasky JA, Sanchez CG. Sirtuin 3 deregulation promotes pulmonary fibrosis. J Gerontol: Ser A. 2016;72(5):595–602. https://doi.org/10.1093/gerona/glw151.
Zhu X, Chu H, Jiang S, Liu Q, Liu L, Xue Y, et al. Sirt1 ameliorates systemic sclerosis by targeting the mTOR pathway. J Dermatol Sci. 2017;87(2):149–58. https://doi.org/10.1016/j.jdermsci.2017.04.013.
Bindu S, Pillai VB, Kanwal A, Samant S, Mutlu GM, Verdin E, et al. SIRT3 blocks myofibroblast differentiation and pulmonary fibrosis by preventing mitochondrial DNA damage. Am J Phys Lung Cell Mol Phys. 2017;312(1):L68–78. https://doi.org/10.1152/ajplung.00188.2016.
Jafarinejad-Farsangi S, Farazmand A, Gharibdoost F, Karimizadeh E, Noorbakhsh F, Faridani H, et al. Inhibition of microRNA-21 induces apoptosis in dermal fibroblasts of patients with systemic sclerosis. Int J Dermatol. 2016;55(11):1259–67. https://doi.org/10.1111/ijd.13308.
Wermuth PJ, Sonsoles PV, Jimenez SA. Exosomes isolated from serum of systemic sclerosis patients display alterations in their content of profibrotic and antifibrotic microRNA and induce a profibrotic phenotype in cultured normal dermal fibroblasts. Clin Exp Rheumatol. 2017.
Dolcino M, Pelosi A, Fiore PF, Patuzzo G, Tinazzi E, Lunardi C, et al. Gene profiling in patients with systemic sclerosis reveals the presence of oncogenic gene signatures. Front Immunol. 2018;9:449. https://doi.org/10.3389/fimmu.2018.00449.
Chouri E, Servaas NH, Bekker CPJ, Affandi AJ, Cossu M, Hillen MR, et al. Serum microRNA screening and functional studies reveal miR-483-5p as a potential driver of fibrosis in systemic sclerosis. J Autoimmun. 2018;89:162–70. https://doi.org/10.1016/j.jaut.2017.12.015.
Tsou PS, Khanna D, Sawalha AH. Identifying CYR61 as a potential anti-fibrotic and pro-angiogenic mediator in scleroderma. Arthritis Rheum. 2019. https://doi.org/10.1002/art.40890.
Palumbo-Zerr K, Zerr P, Distler A, Fliehr J, Mancuso R, Huang J, et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-beta signaling and fibrosis. Nat Med. 2015;21(2):150–8. https://doi.org/10.1038/nm.3777.
• Gallant-Behm CL, Piper J, Lynch JM, Seto AG, Hong SJ, Mustoe TA, et al. A microRNA-29 mimic (Remlarsen) represses extracellular matrix expression and fibroplasia in the skin. J Invest Dermatol. 2019;139(5):1073–81. https://doi.org/10.1016/j.jid.2018.11.007. Examined the pharmacodynamic endpoints and clinical efficacy of a miRNA mimic in humans.
Sun X, Xiao Y, Zeng Z, Shi Y, Tang B, Long H, et al. All-trans retinoic acid induces CD4+CD25+FOXP3+ regulatory T cells by increasing FOXP3 demethylation in systemic sclerosis CD4+ T Cells. J Immunol Res. 2018;2018:8658156. https://doi.org/10.1155/2018/8658156.
Bujor AM, Haines P, Padilla C, Christmann RB, Junie M, Sampaio-Barros PD, et al. Ciprofloxacin has antifibrotic effects in scleroderma fibroblasts via downregulation of Dnmt1 and upregulation of Fli1. Int J Mol Med. 2012;30(6):1473–80. https://doi.org/10.3892/ijmm.2012.1150.
Brueckner B, Rius M, Markelova MR, Fichtner I, Hals PA, Sandvold ML, et al. Delivery of 5-azacytidine to human cancer cells by elaidic acid esterification increases therapeutic drug efficacy. Mol Cancer Ther. 2010;9(5):1256–64. https://doi.org/10.1158/1535-7163.MCT-09-1202.
Naz A, Cui Y, Collins CJ, Thompson DH, Irudayaraj J. PLGA-PEG nano-delivery system for epigenetic therapy. Biomed Pharmacother. 2017;90:586–97. https://doi.org/10.1016/j.biopha.2017.03.093.
Jeffries MA. Epigenetic editing: how cutting-edge targeted epigenetic modification might provide novel avenues for autoimmune disease therapy. Clin Immunol. 2018;196:49–58. https://doi.org/10.1016/j.clim.2018.02.001.
Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. https://doi.org/10.1186/gb-2013-14-10-r115.
Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–67. https://doi.org/10.1016/j.molcel.2012.10.016.
Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17(1):171. https://doi.org/10.1186/s13059-016-1030-0.
Horvath S, Ritz BR. Increased epigenetic age and granulocyte counts in the blood of Parkinson's disease patients. Aging (Albany NY). 2015;7(12):1130–42. https://doi.org/10.18632/aging.100859.
Dugue PA, Bassett JK, Joo JE, Jung CH, Ming Wong E, Moreno-Betancur M, et al. DNA methylation-based biological aging and cancer risk and survival: pooled analysis of seven prospective studies. Int J Cancer. 2018;142(8):1611–9. https://doi.org/10.1002/ijc.31189.
Vidal-Bralo L, Lopez-Golan Y, Mera-Varela A, Rego-Perez I, Horvath S, Zhang Y, et al. Specific premature epigenetic aging of cartilage in osteoarthritis. Aging. 2016;8(9):2222–31. https://doi.org/10.18632/aging.101053.
Dozmorov MG, Coit P, Maksimowicz-McKinnon K, Sawalha AH. Age-associated DNA methylation changes in naive CD4(+) T cells suggest an evolving autoimmune epigenotype in aging T cells. Epigenomics. 2017;9(4):429–45. https://doi.org/10.2217/epi-2016-0143.
Acknowledgments
This work was supported by MICHR grant UL1TR002240, the Scleroderma Foundation, the Arthritis National Research Foundation, the American Autoimmune Related Disease Foundation, the Edward D. and Ellen K. Dryer Charitable Foundation, Dr. Donna Shelley, and Mr. Lawrence Shelley, as well as Mr. Craig Sincock and Mrs. Sue Sincock.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The author declares that she has no potential conflicts of interest relevant to this article.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by the author.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Scleroderma
Rights and permissions
About this article
Cite this article
Tsou, PS. Epigenetic Control of Scleroderma: Current Knowledge and Future Perspectives. Curr Rheumatol Rep 21, 69 (2019). https://doi.org/10.1007/s11926-019-0877-y
Published:
DOI: https://doi.org/10.1007/s11926-019-0877-y