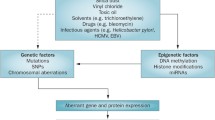
Abstract
Systemic sclerosis (SSc) is an autoimmune connective tissue disease characterized by vascular injury, activation of the immune system, and diffuse tissue fibrosis. The precise etiology of SSc is undetermined, but there is evidence suggestive of a connection between environmental factors and SSc pathogenesis. In general, harmful environmental factors are sensed by the epigenetic regulatory mechanisms that alter host gene expression leading to the emergence of disease-specific phenotype. There are three epigenetic mechanisms involved in gene regulation: DNA methylation, histone modifications, and microRNAs. Although there is evidence that SSc phenotype could be, to a some degree, determined by genetic variants, it is clear now that non-genetic factors outweigh the genetic risk in SSc. Accordingly, the environment can trigger epigenetic regulation that in turn establishes a molecular framework linking environmental exposures to genetics, leading to the disease process, possibly in a genetically predisposed host. Although we have just begun to appreciate the potential role of epigenetics in SSc, many important and promising clues have been observed. In this review, we will summarize the work that has been done in the field of epigenetic regulation in SSc, and we will discuss possible factors and mechanisms that may lead to epigenetic dysregulation in SSc.


Similar content being viewed by others
References
Abraham DJ, Varga J (2005) Scleroderma: from cell and molecular mechanisms to disease models. Trends Immunol 26(11):587–595
Feghali-Bostwick C, Medsger TA Jr, Wright TM (2003) Analysis of systemic sclerosis in twins reveals low concordance for disease and high concordance for the presence of antinuclear antibodies. Arthritis Rheum 48(7):1956–1963
Valesini G et al (1993) Geographical clustering of scleroderma in a rural area in the province of Rome. Clin Exp Rheumatol 11(1):41–47
Silman AJ et al (1990) Geographical clustering of scleroderma in south and west London. Br J Rheumatol 29(2):93–96
Russo VEA, Martienssen RA, Riggs AD (1996) Epigenetic mechanisms of gene regulation. Cold Spring Harbor monograph series. Cold Spring Harbor Laboratory Press, Plainview, p 692
Szyf M (2015) Nongenetic inheritance and transgenerational epigenetics. Trends Mol Med 21(2):134–144
Plongthongkum N, Diep DH, Zhang K (2014) Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat Rev Genet 15(10):647–661
Okano M et al (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99(3):247–257
Comb M, Goodman HM (1990) CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res 18(13):3975–3982
Lewis JD et al (1992) Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69(6):905–914
Ito S et al (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466(7310):1129–1133
Altorok N et al (2014) Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann Rheum Dis
Wang Y, Fan PS, Kahaleh B (2006) Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum 54(7):2271–2279
Egging D et al (2007) Wound healing in tenascin-X deficient mice suggests that tenascin-X is involved in matrix maturation rather than matrix deposition. Connect Tissue Res 48(2):93–98
Kubo M et al (2003) Persistent down-regulation of Fli1, a suppressor of collagen transcription, in fibrotic scleroderma skin. Am J Pathol 163(2):571–581
Kimura A et al (2010) Runx1 and Runx2 cooperate during sternal morphogenesis. Development 137(7):1159–1167
Zhao Q et al (1997) Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn 209(4):377–386
Yoshida CA et al (2004) Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev 18(8):952–963
Ihn H (2008) Autocrine TGF-beta signaling in the pathogenesis of systemic sclerosis. J Dermatol Sci 49(2):103–113
Blobe GC, Schiemann WP, Lodish HF (2000) Role of transforming growth factor beta in human disease. N Engl J Med 342(18):1350–1358
Margadant C, Sonnenberg A (2010) Integrin-TGF-beta crosstalk in fibrosis, cancer and wound healing. EMBO Rep 11(2):97–105
Asano Y et al (2005) Increased expression of integrin alpha(v)beta3 contributes to the establishment of autocrine TGF-beta signaling in scleroderma fibroblasts. J Immunol 175(11):7708–7718
Asano Y et al (2006) Increased expression of integrin alphavbeta5 induces the myofibroblastic differentiation of dermal fibroblasts. Am J Pathol 168(2):499–510
Asano Y et al (2004) Increased expression levels of integrin alphavbeta5 on scleroderma fibroblasts. Am J Pathol 164(4):1275–1292
Horan GS et al (2008) Partial inhibition of integrin alpha(v)beta6 prevents pulmonary fibrosis without exacerbating inflammation. Am J Respir Crit Care Med 177(1):56–65
Shi-Wen X et al (2007) Endogenous endothelin-1 signaling contributes to type I collagen and CCN2 overexpression in fibrotic fibroblasts. Matrix Biol 26(8):625–632
Atfi A et al (2007) The disintegrin and metalloproteinase ADAM12 contributes to TGF-beta signaling through interaction with the type II receptor. J Cell Biol 178(2):201–208
Skubitz KM, Skubitz AP (2004) Gene expression in aggressive fibromatosis. J Lab Clin Med 143(2):89–98
Taniguchi T et al (2013) Serum levels of ADAM12-S: possible association with the initiation and progression of dermal fibrosis and interstitial lung disease in patients with systemic sclerosis. J Eur Acad Dermatol Venereol 27(6):747–753
Wei J et al (2011) Canonical Wnt signaling induces skin fibrosis and subcutaneous lipoatrophy: a novel mouse model for scleroderma? Arthritis Rheum 63(6):1707–1717
Lam AP et al (2011) Nuclear beta-catenin is increased in systemic sclerosis pulmonary fibrosis and promotes lung fibroblast migration and proliferation. Am J Respir Cell Mol Biol 45(5):915–922
Dees C et al (2013) The Wnt antagonists DKK1 and SFRP1 are downregulated by promoter hypermethylation in systemic sclerosis. Ann Rheum Dis
Altorok N, Wang Y, Kahaleh B (2014) Endothelial dysfunction in systemic sclerosis. Curr Opin Rheumatol 26(6):615–620
Romero LI et al (2000) Differential expression of nitric oxide by dermal microvascular endothelial cells from patients with scleroderma. Vasc Med 5(3):147–158
Fish JE, Marsden PA (2006) Endothelial nitric oxide synthase: insight into cell-specific gene regulation in the vascular endothelium. Cell Mol Life Sci 63(2):144–162
Wang Y, KB (2007) Epigenetic regulation in scleroderma: high-throughput DNA methylation profiling of Ssc fibroblasts and microvascular endothelial cells and the central role for Nos3 and Fli1 epigenetic repression in the emergence of Ssc cellular phenotype [abstract]. American College of Rheumatology; Annual scientific meeting.
Sgonc R et al (1996) Endothelial cell apoptosis is a primary pathogenetic event underlying skin lesions in avian and human scleroderma. J Clin Invest 98(3):785–792
Wang,Y, Kahaleh B (2013) Epigenetic repression of bone morphogenetic protein receptor II expression in scleroderma. J Cell Mol Med
Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190:372–373
Lian X et al (2012) DNA demethylation of CD40l in CD4+ T cells from women with systemic sclerosis: a possible explanation for female susceptibility. Arthritis Rheum 64(7):2338–2345
Lu Q et al (2007) Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol 179(9):6352–6358
Uz E et al (2008) Skewed X-chromosome inactivation in scleroderma. Clin Rev Allergy Immunol 34(3):352–355
Denoeud J, Moser M (2011) Role of CD27/CD70 pathway of activation in immunity and tolerance. J Leukoc Biol 89(2):195–203
Jiang H et al (2012) Demethylation of TNFSF7 contributes to CD70 overexpression in CD4+ T cells from patients with systemic sclerosis. Clin Immunol 143(1):39–44
Stummvoll GH et al (2004) Increased transendothelial migration of scleroderma lymphocytes. Ann Rheum Dis 63(5):569–574
Wang Y et al (2014) Hypomethylation and overexpression of ITGAL (CD11a) in CD4(+) T cells in systemic sclerosis. Clin Epigenetics 6(1):25
Kramer M et al (2013) Inhibition of H3K27 histone trimethylation activates fibroblasts and induces fibrosis. Ann Rheum Dis 72(4):614–620
Glazer RI et al (1986) 3-Deazaneplanocin: a new and potent inhibitor of S-adenosylhomocysteine hydrolase and its effects on human promyelocytic leukemia cell line HL-60. Biochem Biophys Res Commun 135(2):688–694
Wang Y et al (2013) Aberrant histone modification in peripheral blood B cells from patients with systemic sclerosis. Clin Immunol 149(1):46–54
Nilsen TW (2007) Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet 23(5):243–249
van Rooij E et al (2008) Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A 105(35):13027–13032
Liu Y et al (2010) Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension 55(4):974–982
Pandit KV, Milosevic J, Kaminski N (2011) MicroRNAs in idiopathic pulmonary fibrosis. Transl Res 157(4):191–199
Kriegel AJ et al (2012) The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics 44(4):237–244
Maurer B et al (2010) MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum 62(6):1733–1743
Zhu H et al (2012) MicroRNA expression abnormalities in limited cutaneous scleroderma and diffuse cutaneous scleroderma. J Clin Immunol 32(3):514–522
Zhu H et al (2013) MicroRNA-21 in scleroderma fibrosis and its function in TGF-beta-regulated fibrosis-related genes expression. J Clin Immunol 33(6):1100–1109
Sing T et al (2012) microRNA-92a expression in the sera and dermal fibroblasts increases in patients with scleroderma. Rheumatology (Oxford) 51(9):1550–1556
Honda N et al (2012) TGF-beta-mediated downregulation of microRNA-196a contributes to the constitutive upregulated type I collagen expression in scleroderma dermal fibroblasts. J Immunol 188(7):3323–3331
Wang Y, Kahaly O, Kahaleh B (2010) Down-regulated microRNA-152 induces aberrant DNA methylation in scleroderma endothelial cells by targeting DNA methyltransferase 1. [abstract]. Arthritis Rheum 62(Suppl 10):1352
Torrens C, Poston L, Hanson MA (2008) Transmission of raised blood pressure and endothelial dysfunction to the F2 generation induced by maternal protein restriction in the F0, in the absence of dietary challenge in the F1 generation. Br J Nutr 100(4):760–766
Altorok N et al (2014) Epigenetics, the holy grail in the pathogenesis of systemic sclerosis. Rheumatology (Oxford)
Rodnan GP et al (1967) The association of progressive systemic sclerosis (scleroderma) with coal miners’ pneumoconiosis and other forms of silicosis. Ann Intern Med 66(2):323–334
Yamakage A et al (1980) Occupational scleroderma-like disorder occurring in men engaged in the polymerization of epoxy resins. Dermatologica 161(1):33–44
Czirjak L, Szegedi G (1987) Benzene exposure and systemic sclerosis. Ann Intern Med 107(1):118
Owens GR, Medsger TA (1988) Systemic sclerosis secondary to occupational exposure. Am J Med 85(1):114–116
Rakyan VK et al (2011) Epigenome-wide association studies for common human diseases. Nat Rev Genet 12(8):529–541
Lorthongpanich C et al (2013) Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science 341(6150):1110–1112
Ficz G et al (2011) Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473(7347):398–402
Chia N et al (2011) Hypothesis: environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics 6(7):853–856
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is a contribution to the Special Issue on Immunopathology of Systemic Sclerosis - Guest Editors: Jacob M. van Laar and John Varga
Rights and permissions
About this article
Cite this article
Altorok, N., Kahaleh, B. Epigenetics and systemic sclerosis. Semin Immunopathol 37, 453–462 (2015). https://doi.org/10.1007/s00281-015-0504-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00281-015-0504-6