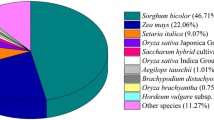
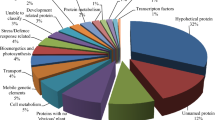
Abstract
Jerusalem artichoke (Helianthus tuberosus L.), a plant of the Asteraceae family, is widely cultivated for its multiple pharmacological properties and is being developed as a renewable feedstock, as well as a source of biofuels and biochemicals for industrial applications. Despite its nutritional benefits and economic potential, transcriptomic and genomic information is scarce. In the present study, we performed phenotypic characterization and RNA-Seq analysis of three Jerusalem artichoke cultivars, “Purple Jerusalem artichoke” (PJA), “Hindung Jerusalem artichoke” (HJA) and “Dafeng Jerusalem artichoke” (DJA). The cultivars exhibited obvious differences in their responses to drought, high salinity and oxidative stress, as well as morphological variations. The PJA cultivar had the highest concentration of anthocyanin, and the DJA cultivar had the strongest tolerance to environmental stresses among the three cultivars. Based on the three assembled transcriptomes, we identified 2435, 3283 and 3657 putative cultivar-specific transcripts from leaf and tuber tissues in cultivars PJA, HJA and DJA, respectively, which enlarges the pool of transcriptomes available for Jerusalem artichoke. We also detected 11,319, 13,190 and 12,717 potential cultivar-specific simple sequence repeats (SSRs) from the transcriptomic data for, PJA, HJA and DJA, respectively. In addition, five SSRs were identified as candidate molecular markers for cultivar identification, as determined by genomic PCR analysis. Our comparative RNA-Seq analysis and de novo transcriptome assemblies constitute a comprehensive transcriptome resource and provide essential sequence information for identifying Jerusalem artichoke cultivars. These results should therefore be useful for future gene discovery, molecular studies and agricultural improvement of this important non-model species.





Similar content being viewed by others
References
Bal LK (1997) Performance of several Jerusalem artichoke clones (Helianthus tuberosus L.) screened for adaptability in Korea. J Korean Soc Grassl Sci 17(3):305–314
Barakat A, DiLoreto DS, Zhang Y, Smith C, Baier K, Powell WA, Wheeler N, Sederoff R, Carlson JE (2009) Comparison of the transcriptomes of American chestnut (Castanea dentata) and Chinese chestnut (Castanea mollissima) in response to the chestnut blight infection. BMC Plant Biol 9:51
Chi ZM, Zhang T, Cao TS, Liu XY, Cui W, Zhao CH (2011) Biotechnological potential of inulin for bioprocesses. Bioresour Technol 102:4295–4303
Cochrane FC, Davin LB, Lewis NG (2004) The Arabidopsis phenylalanine ammonia lyase gene family: kinetic characterization of the four PAL isoforms. Phytochemistry 65:1557–1564
Conesa A, Gotz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008:619832
Dixon DP, Sellars JD, Edwards R (2011) The Arabidopsis phi class glutathione transferase AtGSTF2: binding and regulation by biologically active heterocyclic ligands. Biochem J 438:63–70
Dorn KM, Fankhauser JD, Wyse DL, Marks MD (2013) De novo assembly of the pennycress (Thlaspi arvense) transcriptome provides tools for the development of a winter cover crop and biodiesel feedstock. Plant J 75:1028–1038
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucl Acids Res 38:W64–W70
Ferreira de Carvalho J, Poulain J, Da Silva C, Wincker P, Michon-Coudouel S, Dheilly A, Naquin D, Boutte J, Salmon A, Ainouche M (2013) Transcriptome de novo assembly from next-generation sequencing and comparative analyses in the hexaploid salt marsh species Spartina maritima and Spartina alterniflora (Poaceae) Heredity (Edinb) 110:181–193
Firon N, LaBonte D, Villordon A, Kfir Y, Solis J, Lapis E, Perlman TS, Doron-Faigenboim A, Hetzroni A, Althan L, Adani Nadir L (2013) Transcriptional profiling of sweetpotato (Ipomoea batatas) roots indicates down-regulation of lignin biosynthesis and up-regulation of starch biosynthesis at an early stage of storage root formation. BMC Genom 14:460
Fuchs A (2012) Inulin and inulin-containing crops. Elsevier, Berlin
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012) Phytozome: a comparative platform for green plant genomics. Nucl Acids Res 40:D1178–D1186
Iseli C, Jongeneel CV, Bucher P (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. In: Proceedings of the international conference on intelligent systems for molecular biology, pp 138–148
Jia HM, Shen YT, Jiao Y, Wang GY, Dong X, Jia HJ, Du F, Liang SM, Zhou CC, Mao WH, Gao ZS (2014) Development of 107 SSR markers from whole genome shotgun sequences of Chinese bayberry (Myrica rubra) and their application in seedling identification. J Zhejiang Univ Sci B 15:997–1005
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240
Jung WY, Lee SS, Kim CW, Kim HS, Min SR, Moon JS, Kwon SY, Jeon JH, Cho HS (2014) RNA-seq analysis and de novo transcriptome assembly of Jerusalem artichoke (Helianthus tuberosus Linne). PLoS ONE 9:e111982
Kaur N, Gupta AK (2002) Applications of inulin and oligofructose in health and nutrition. J Biosci 27:703–714
Kaur S, Cogan NO, Pembleton LW, Shinozuka M, Savin KW, Materne M, Forster JW (2011) Transcriptome sequencing of lentil based on second-generation technology permits large-scale unigene assembly and SSR marker discovery. BMC Genom 12:265
Kays SJ, Nottingham SF (2007) Biology and chemistry of Jerusalem artichoke: Helianthus tuberosus L. CRC press, USA
Kim HA, Lim CJ, Kim S, Choe JK, Jo SH, Baek N, Kwon SY (2014) High-throughput sequencing and de novo assembly of Brassica oleracea var. Capitata L. for transcriptome analysis. PLoS One 9:e92087
Kubo H, Peeters AJ, Aarts MG, Pereira A, Koornneef M (1999) ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 11:1217–1226
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Li L, Li L, Wang Y, Du Y, Qin S (2013) Biorefinery products from the inulin-containing crop Jerusalem artichoke. Biotechnol Lett 35:471–477
Mandel JR, Dechaine JM, Marek LF, Burke JM (2011) Genetic diversity and population structure in cultivated sunflower and a comparison to its wild progenitor, Helianthus annuus L. Theor Appl Genet 123:693–704
Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18:1509–1517
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucl Acids Res 35:W182–W185
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Muthusamy M, Uma S, Backiyarani S, Saraswathi MS (2015) Genome-wide screening for novel, drought stress-responsive long non-coding RNAs in drought-stressed leaf transcriptome of drought-tolerant and -susceptible banana (Musa spp) cultivars using Illumina high-throughput sequencing. Plant Biotechnol Rep 9:279–286
Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344–1349
Novaes E, Drost DR, Farmerie WG, Pappas GJ Jr, Grattapaglia D, Sederoff RR, Kirst M (2008) High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genom 9:312
Ohl S, Hedrick SA, Chory J, Lamb CJ (1990) Functional properties of a phenylalanine ammonia-lyase promoter from Arabidopsis. Plant Cell 2:837–848
Panchev I, Delchev N, Kovacheva D, Slavov A (2011) Physicochemical characteristics of inulins obtained from Jerusalem artichoke (Helianthus tuberosus L.) Eur Food Res Technol 233:889–896
Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta (BBA) Bioenerg 975:384–394
Prasath D, Karthika R, Habeeba NT, Suraby EJ, Rosana OB, Shaji A, Eapen SJ, Deshpande U, Anandaraj M (2014) Comparison of the transcriptomes of ginger (Zingiber officinale Rosc.) and mango ginger (Curcuma amada Roxb.) in response to the bacterial wilt infection. PLoS ONE 9:e99731
Praznik W, Cieslik E, Filipiak-Florkiewicz A (2002) Soluble dietary fibres in Jerusalem artichoke powders: composition and application in bread. Nahrung 46:151–157
Riggins CW, Peng Y, Stewart CN Jr, Tranel PJ (2010) Characterization of de novo transcriptome for waterhemp (Amaranthus tuberculatus) using GS-FLX 454 pyrosequencing and its application for studies of herbicide target-site genes. Pest Manag Sci 66:1042–1052
Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Haweker H, Dong X, Robatzek S, Schulze-Lefert P (2009) Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J 28:3439–3449
Sanchez OJ, Cardona CA (2008) Trends in biotechnological production of fuel ethanol from different feedstocks. Bioresour Technol 99:5270–5295
Schulz MH, Zerbino DR, Vingron M, Birney E (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28:1086–1092
Shin S-Y, Shin C (2016) Regulatory non-coding RNAs in plants: potential gene resources for the improvement of agricultural traits. Plant Biotechnol Rep 10:35–47
Strack DWV (1989) Anthocyanins. Methods Plant Biol 1:325–356
Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA (2003) The COG database: an updated version includes eukaryotes. BMC Bioinform 4:41
Ueno S, Moriguchi Y, Uchiyama K, Ujino-Ihara T, Futamura N, Sakurai T, Shinohara K, Tsumura Y (2012) A second generation framework for the analysis of microsatellites in expressed sequence tags and the development of EST-SSR markers for a conifer, Cryptomeria japonica. BMC Genom 13:136
van der Meer IM, Koops AJ, Hakkert JC, van Tunen AJ (1998) Cloning of the fructan biosynthesis pathway of Jerusalem artichoke. Plant J 15:489–500
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Victoria FC, da Maia LC, de Oliveira AC (2011) In silico comparative analysis of SSR markers in plants. BMC Plant Biol 11:15
Wang S, Wang X, He Q, Liu X, Xu W, Li L, Gao J, Wang F (2012) Transcriptome analysis of the roots at early and late seedling stages using Illumina paired-end sequencing and development of EST-SSR markers in radish. Plant Cell Rep 31:1437–1447
Wang Z, Yu G, Shi B, Wang X, Qiang H, Gao H (2014) Development and characterization of simple sequence repeat (SSR) markers based on RNA-sequencing of Medicago sativa and in silico mapping onto the M. truncatula genome. PLoS ONE 9:e92029
Ward JA, Ponnala L, Weber CA (2012) Strategies for transcriptome analysis in nonmodel plants. Am J Bot 99:267–276
Wei W, Qi X, Wang L, Zhang Y, Hua W, Li D, Lv H, Zhang X (2011) Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genom 12:451
Wei L, Li S, Liu S, He A, Wang D, Wang J, Tang Y, Wu X (2014) Transcriptome analysis of Houttuynia cordata Thunb. by Illumina paired-end RNA sequencing and SSR marker discovery. PLoS ONE 9:e84105
Wei C, Tao X, Li M, He B, Yan L, Tan X, Zhang Y (2015) De novo transcriptome assembly of Ipomoea nil using Illumina sequencing for gene discovery and SSR marker identification. Mol Genet Genomics 290:1873–1884
Wu G, Zhang L, Yin Y, Wu J, Yu L, Zhou Y, Li M (2015) Sequencing, de novo assembly and comparative analysis of Raphanus sativus transcriptome Front. Plant Sci 6:198
Xue YF, Liu ZP (2008) Antioxidant enzymes and physiological characteristics in two Jerusalem artichoke cultivars under salt stress Russ. J Plant Physiol 55:776–781
Zeid M, Mitchell S, Link W, Carter M, Nawar A, Fulton T, Kresovich S (2009) Simple sequence repeats (SSRs) in faba bean: new loci from Orobanche-resistant cultivar ‘Giza 402’. Plant Breeding 128:149–155
Zhang LY, Bernard M, Leroy P, Feuillet C, Sourdille P (2005) High transferability of bread wheat EST-derived SSRs to other cereals. Theor Appl Genet 111:677–687
Zhang H, Wei L, Miao H, Zhang T, Wang C (2012) Development and validation of genic-SSR markers in sesame by RNA-seq. BMC Genom 13:316
Acknowledgements
This work was supported by the Agricultural Biotechnology Developmental Program (Nos. 114061-3, 115076-2 and 116091-3) Grants from the Ministry of Agriculture, Food and Rural Affairs and KRIBB Research Initiative Program to HS Cho.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
11816_2016_421_MOESM4_ESM.tif
Figure S1. Coverage distribution of H. tuberosus transcripts against integrated representative transcripts, as revealed by BLAST analysis. a Comparison of integrated RTs and H. tuberosus loci. b Comparison of integrated RTs and H. tuberosus ESTs using sequence similarity searches. (TIFF 588 kb)
11816_2016_421_MOESM5_ESM.tif
Figure S2. Length distribution of assembled representative transcripts from the transcriptomes of the three cultivars. A comparison of the distribution of assembled representative transcript lengths from the three cultivars is shown. The length of the assembled transcripts vs. number of transcripts is shown. (TIFF 178 kb)
11816_2016_421_MOESM6_ESM.tif
Figure S3. Validation of cultivar-specific gene expression in the three cultivars. a The expression patterns of cultivar-specific genes from the three cultivars are shown. Total RNAs from three cultivars of leaves and tubers were analyzed using RT-PCR. b Expression of anthocyanin-related genes in the tubers of the three cultivars. Expression of HtActin7 was used an internal standard. An aliquot of each RT-PCR mixture was electrophoresed on a 1% agarose gel and visualized by ethidium bromide staining. (TIFF 534 kb)
Rights and permissions
About this article
Cite this article
Jung, W.Y., Lee, S.S., Park, H.J. et al. Comparative transcriptome profiling and SSR marker identification in three Jerusalem artichoke (Helianthus tuberosus L.) cultivars exhibiting phenotypic variation. Plant Biotechnol Rep 10, 447–461 (2016). https://doi.org/10.1007/s11816-016-0421-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-016-0421-8