Abstract
There are several important factors affecting the rate of starch decomposition in plants, including the circadian clock, the regulation of gene expression, the regulation of enzyme activities and starch phosphorylation by glucan, water dikinase activities (GWDs). One isoform of glucan, water dikinase named GWD3 or PWD (EC 2.7.9.5) was isolated for the first time from Arabidopsis thaliana, and now we report its isolation and identification in Solanum tuberosum L. leaves and tubers. We compare StGWD3 sequence to the other GWDs sequences using bioinformatics tools and propose also structural models for the starch-binding domains in StGWD3 and StGWD1. The StGWD3 gene expression and protein were localised in different heterotrophic and autotrophic potato tissues and organs using in situ RT-PCR and immunolocalisation methods, respectively. Diurnal changes in the transcript abundance of StGWD3 in leaves were analysed using quantitative real-time PCR and they appeared to be typical for most genes involved in starch degradation in chloroplasts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Starch is the main storage carbohydrate in higher plants. It is present in all tissues and plays an important role both as a main ingredient in many human diets and as a raw material in many food and chemical industries. Starch metabolism is very important for understanding the biology of the regulation of mass increase in plants. Starch is a complex polysaccharide with two distinct forms, amylose (linear) and amylopectin (branched), which together constitute the starch granule. One of the covalent modifications of starch granules is the phosphoesterification of the C-3 or C-6 hydroxyl group in the glucosyl unit. In storage starch of potato tubers, approximately 0.5 % of the glucose residues is phosphorylated (Mikkelsen et al. 2006), whereas the level of phosphorylation in transitory starch does not exceed 0.1 %. The phosphorylation of starch granules possibly facilitates the attack of amylases on the granule. Because the structure of the biological starch molecule is complex, the initiation of starch degradation requires structural changes of the starch granule (Kötting et al. 2010; Smith 2012). Phosphorylation of the starch polysaccharide chains can weaken the compact granule and create space for amylases to bind the polysaccharides (Fettke et al. 2006; Dudkiewicz et al. 2008; Hejazi et al. 2008). As suggested by systematic analysis of NMR and Molecular Dynamics Simulation (MDS) data for five amylopectin branch-point trisaccharides (Hansen et al. 2008), there are several inter-ring bridging water molecules involved in the stabilisation of specific amylopectin conformations within the starch granule. The introduction of additional negatively charged phosphate groups could possibly change the hydration network between oxygen atoms in monosaccharide rings, which could influence the structure of the entire granule. According to recently published NMR and MDS results from Hansen et al. (2009), 3-O-phosphorylation of disaccharides strongly influences the glucosidic bonds in starch. The observed shift in the conformational equilibrium was induced only by 3-O-phosphorylation and has been attributed to the steric effects of the introduced phosphate group. The frequency of phosphorylated glucose residues in storage starch in potato tubers is 1 per 200–300 (Mikkelsen et al. 2004). Phosphorylation of starch occurs widely in many plant species, but the level of phosphoesterification depends on the botanical origin of starch granules. The proposed mechanism of starch phosphorylation due to StGWD and AtGWD was recently published (Hejazi et al. 2012). This process seems to depend strongly on availability of the various acceptors during the phosphotransfer reaction catalysed by GWDs.
Glucan, water dikinase (GWD1; EC 2.7.9.4), known as R1 protein in S. tuberosum and SEX1 in A. thaliana, phosphorylates glucose residues at the C-6 position (Ritte et al. 2006). The second enzyme, the GWD3, named also phosphoglucan, water dikinase (PWD/GWD3; EC 2.7.9.5) (Baunsgaard et al. 2005; Kötting et al. 2005; Ritte et al. 2006; Smith 2012), phosphorylates the C-3 position of glucose residues in amylopectin chains. Both dikinases are necessary for normal starch metabolism in Arabidopsis (Lloyd et al. 2005; Fettke et al. 2006; Edner et al. 2007). GWD3 (PWD) activity depends highly on GWD1 action because the former phosphorylates substrates that have already been phosphorylated, such glucans have proper structure (Kötting et al. 2005, Hejazi et al. 2009) and therefore acts downstream of GWD1.
Three genes have been identified in the Arabidopsis genome that encodes GWD homologs: GWD1 (GWD, SEX1), GWD2 and GWD3 (PWD). A plastidial localisation has been confirmed for GWD1 and GWD3, but the third homologue, GWD2, has been localised in the cytosol, which excludes its involvement in starch degradation (Orzechowski 2008).
To date, there has been one GWD homologue, previously called R1, described in potato. Its activity depends on the cellular redox potential (Mikkelsen et al. 2004, 2005, 2006). After analysing potato EST sequences, Mikkelsen et al. (2005) suggested that there is at least one additional GWD homologue in S. tuberosum that is independent of the redox potential, similar to GWD3 in Arabidopsis.
In this paper, we describe the purification of the second GWD homologue, named StGWD3, from potato tubers, its identification using MALDI-TOF, analysis of expression and the localisation of its gene and protein using in situ RT-PCR or polyclonal antibody labelling, respectively. Using two-step real-time RT-PCR, we analysed diurnal changes in StGWD3 and StGWD1 transcript levels. We present the full nucleotide sequence of StGWD3 and the results of the bioinformatics analysis of its domain structure and substrate-binding site. We compared the effects of phosphorylation on intermolecular energies (kJ/mol) for stabilising complexes between the set of sugar ligands and S. tuberosum starch-binding domains (SBDs) from the StGWD1 and StGWD3 enzymes. We observed that phosphorylation of the oligosaccharides affects ligand binding to the SBDs in both StGWD1 and StGWD3. As Glaring et al. (2007) used the term “GWD2” for the cytosolic homologue of GWD1 in Arabidopsis, we suggest that this novel GWD homologue be named “StGWD3” to avoid confusion in the terminology given by previous works (Baunsgaard et al. 2005; Kötting et al. 2005; Orzechowski 2008; Blennow and Engelsen 2010).
Materials and methods
Potato Solanum tuberosum L. cv. “Russet Burbank” plants were grown under controlled conditions in a growth chamber at 23 °C and 60 % relative humidity under a 16-h photoperiod (100 μmol quanta m−2 s−1). Harvested leaves and tubers, which were stored in the dark either at room temperature or at 4 °C (cold-stored for 8 weeks), were collected and stored at −80 °C before analysis.
Purification of StGWD3
All purification steps were performed at 4 °C. Approximately 100 g potato tubers were ground in extraction buffer (100 mM citrate, 20 mM Tris, 2 μM leupeptin hemisulfate, 2 μM pepstatin A, 1 mM EDTA, 5 mM 2-mercaptoethanol, pH 6.5) using 2 ml buffer per 1 g tubers. The homogenate was filtered through two layers of gauze and then centrifuged for 20 min at 14,850g. The pellet was discarded, and the supernatant was precipitated using 30 % ammonium sulphate with stirring for approximately 20 min. The solution was centrifuged again for 20 min, and the supernatant was precipitated with 60 % ammonium sulphate and treated as before. After centrifugation, the pellet was dissolved in 15 ml extraction buffer and centrifuged for 10 min at 14,850g. To desalt the protein fraction following centrifugation, the supernatant was loaded onto a Sephadex G-25 column (1 × 11 cm) equilibrated with 25 ml extraction buffer. A total volume of 2 ml of supernatant was deposited on the column, and 4 ml of eluted extraction buffer was collected as a separate fraction. After collecting each fraction, the column was re-equilibrated. The obtained fractions were then loaded onto an amylose resin (BioLabs) column (1.5 × 2.5 cm; ~5 ml resin) for affinity chromatography and equilibrated with 25 ml extraction buffer. The flow speed was approximately 1 ml min−1. The column was washed with 1:10 diluted extraction buffer, and the flow through was discarded. The column was eluted with 1:10 diluted extraction buffer containing 2 % Dextrin 20 (Fluka). Fractions (1.5 ml) were collected into microcentrifuge tubes. Most of the proteins that specifically bound to the resin were eluted in fractions 2–6. The collected fractions were concentrated to 0.1–0.2 ml in a vacuum centrifuge.
Denaturating electrophoresis
One-dimensional SDS-PAGE was performed according to the protocol of Laemmli (1970). Proteins were stained with Coomassie Blue R-250 (Carl Roth GmbH).
In-gel digestion, mass spectrometry and database search
After in-gel digestion of proteins, MALDI-TOF MS analysis was carried out in the Laboratory of Mass Spectrometry, Institute of Biochemistry and Biophysics, Polish Academy of Sciences (Warsaw, Poland) as previously described by Truszkiewicz and Paszkowski (2005). A database search using the proteolytic peptide masses was performed with the Mascot program (http://www.matrixscience.com/search_form_select_html). The search was done with an assumed peptide mass accuracy of ±0.1 Da, and the database was restricted to proteins with a mass below 200 kDa.
RNA isolation
Total RNA was isolated from potato leaves, which were collected in three replicates at eight time points during a single day, using the NucleoSpin RNA Plant Kit (Macherey-Nagel). RNA concentrations were determined spectrophotometrically by absorption at 260, 230 and 280 nm. The quality of the RNA was examined using electrophoresis on a 1.3 % agarose gel.
cDNA synthesis, cloning and sequencing of full-length StGWD3
First-strand cDNA synthesis, primed with an oligo (dT)12–18 primer, was performed with AMV reverse transcriptase following the manufacturer’s protocol (Promega). Based on the sequence information of the A. thaliana GWD3 gene and ESTs from the potato (EST734844, EST439862), a pair of primers was designed to amplify a 1,380-bp fragment from the StGWD3 gene. The sequences of the forward and reverse primers used in the PCR are respectively StGWD3-F (GGTATATCTGGTTGGAAGCC) and StGWD3-R (GCATCTTTCTGGGAGACAC). PCR was carried out with 35 cycles of 94 °C for 60 s, 55 °C for 30 s and 72 °C for 90 s. PCR was performed in 20-μl reaction mixture containing 20 ng of cDNA, 0.2 μM dNTPs, 0.4 μM each primer, 1× GoTaq buffer, 2 mM MgCl2 and 1.25 U GoTaq DNA polymerase (Promega).
The full-length cDNA was generated using the Rapid Amplification of cDNA Ends (RACE) system (Invitrogen). All reactions were performed essentially according to the manufacturer’s instructions. The RACE-ready first-strand cDNA was prepared from RNA isolated from leaves with random primers or a GeneRacer oligo dT primer and used as a template for 5′ RACE and 3′ RACE, respectively. First-strand synthesis was performed with SuperScript III Reverse Transcriptase (Invitrogen).
The gene-specific primers used for RACE were designed based on the above partial StGWD3 cDNA sequence. PCR amplification for 3′ and 5′ RACE was performed with the RACE-F1 (CAGCAGCCGGACTTTATGATTCAATT), RACE-R1 (TGACGGCAATTGAGGAAGCACT) and RACE-R2 (GCCCGATTTCGCAGAGACGCCAACTTC) primers. PCR was performed with Taq DNA High Fidelity Polymerase (Invitrogen).
Based on the nucleotide sequence of the 5′ and 3′ RACE products, the 5′ and 3′ gene-specific primers StGWD3-full-F (ATGGATTCTATGCATCTGTCACACTGTT) and StGWD3-full-R (TCTCACTGGGGTTGAGGTCGCGATT) were designed for the amplification of the full-length StGWD3 cDNA. PCR was carried out at 94 °C for 60 s, followed by 35 cycles of 94 °C for 60 s, 58 °C for 30 s and 72 °C for 3 min 30 s. PCR was performed in 50-μl reaction mixture that contained 1 μl of first-strand cDNA, 0.2 mM dNTPs, 0.4 μM each primer, 1× High Fidelity PCR Buffer, 2 mM MgSO4 and 1 U Platinum Taq DNA High Fidelity Polymerase (Invitrogen).
In all cloning experiments, PCR products were gel-purified with the DNA Extraction Kit (Fermentas), ligated into pGEM-T Easy (Promega) and amplified in E. coli strain JM109. Plasmids were isolated by a standard alkaline lysis method. DNA sequencing was performed with the ABI Prism BigDye Terminator Cycle Sequencing Kit on an ABI Prism 3730 DNA analyser (Applied Biosystems) at the DNA Sequencing and Oligonucleotide Synthesis Laboratory, Institute of Biochemistry and Biophysics, Polish Academy of Sciences (Warsaw, Poland).
Two-step real-time RT-PCR
One microgram of each RNA sample was reverse transcribed into cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche) and anchored oligo(dT)18 primers specific to the poly(A) tail according to the manufacturer’s instructions. cDNA synthesis from three independent RNA extractions was performed for each time point to obtain biological replicates. Based on the sequence information from GenBank (http://www.ncbi.nlm.nih.gov/) of potato StGWD1 (GI:186886419) and of cloned full-length StGWD3 cDNA (GU045560), two primers were designed StGWD3-F (CAATAGCTATGCGTCGGAAGTG) and StGWD3-R (GCTTTGCATTCCTCGGGCTTC). Relative gene expression was evaluated with LightCycler Software 4.1 using a comparative ratio of the examined gene over the reference gene, which was elongation factor 1-α (EF1-α) amplified using the primers designed by Nicot et al. (2005). Gene-specific primers for RT-PCR for StGWD1 are StGWD1-F (CCCACGATCTTAGTAGCAAA) and StGWD1-R (TTAGCTCCAACCATTTCACT) and for EF1-α, EF1-α-F (ATTGGAAACGGATATGCTCCA) and EF1-α-R (TCCTTACCTGAACGCCTGTCA). Real-time RT-PCR was performed using the LightCycler FastStart DNA Master SYBR Green I Kit (Roche Diagnostics), with 250 nM of forward and reverse primers in a Light Cycler 2.0 device (Roche Diagnostics) according to the manufacturer’s instructions. PCR conditions were as follows: initial denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s, 62 °C for 5 s, and 72 °C for 12 s.
In situ RT-PCR
Tissue localisation of StGWD3 transcripts was performed using the Titan One Tube RT-PCR System (Roche) and PTC-100 (MJ Research) thermal cycler. Samples of potato leaves and tubers in early tuberisation phase and those stored at 4 °C were collected, fixed and embedded in butyl-methyl-methacrylate resin (BMM) under RNase-free conditions following procedure described by Fudali et al. (2008). RT was conducted at 50 °C for 45 min, and PCR consisted of 46 cycles of 94 °C for 45 s, 57 °C for 30 s, and 68 °C for 30 s. The same StGWD3 primer pair was used as for real-time RT-PCR. After removing the RT-PCR mixture, the amplified cDNA was detected using the Fluorescent Antibody Enhancer Set for DIG Detection (Roche) or anti-DIG antibodies conjugated with alkaline phosphatase (Roche). In the latter case, the colour was developed following the procedure described by de Almeida et al. (2001). In control experiments, digoxigenin-labelled dUTP or primary detection antibodies were omitted. Sections were examined under an Olympus AX70 ‘Provis’ (Olympus) light microscope equipped with an Olympus DP50 digital camera and a BX-TFC2 fluorescence filter set.
Immunolocalisation of StGWD3
One-dimensional SDS-PAGE was performed using the Laemmli method (1970). Proteins were transferred to nitrocellulose membranes using a modified procedure of Towbin et al. (1979). A polyclonal antibody anti-StGWD3 against the purified StGWD3 isolated from potato tubers was raised in rabbit by Eurogentec (Searing, Belgium). Immunodetection of StGWD3 was carried out with antiserum diluted 1:500.
For microscopic immunocytochemical localisation of StGWD3, samples of potato tubers stored at room temperature or at 4 °C and leaves illuminated for 3 or 10 h were fixed in 2 % (w/v) paraformaldehyde in MSB buffer (50 mM PIPES, 5 mM EDTA, 5 mM MgSO4, pH 6.9) for 2 h. The samples were then dehydrated in a graded series of ethanol solutions and embedded in LR-White acrylic resin (Fudali et al. 2008). After sectioning on a Leica RM2165 microtome, the 3-μm thick sections were incubated in 2 % (w/v) BSA for 1 h and then incubated with primary anti-StGWD3 antibody diluted 1:100 with PBS for 1 h. The primary antibody was detected using a commercially available goat-anti-rabbit secondary antibody conjugated to the Alexa Fluor488 fluorochrome (Invitrogen). Slides were examined under an Olympus AX70 ‘Provis’ (Olympus) microscope equipped with a BX-TFC2 fluorescence filter set and an Olympus DP50 digital camera. The ultrastructural localisation of StGWD3 was observed on 100-nm thick sections taken on a Leica UCT ultramicrotome and collected on uncoated nickel grids (400 mesh). The immunolocalisation procedure was the same as described above, except the secondary antibody was replaced with a goat-anti-rabbit antibody conjugated to Ø20-nm colloidal gold particles. Microscopic examinations were conducted on an FEI M-268D ‘Morgagni’ (FEI) transmission electron microscope equipped with an SIS ‘Morada’ (SIS) digital camera. In control experiments, the primary anti-StGWD3 antibody was omitted.
Homology modelling and sequence alignment
Multiple sequence alignments (MSA) of A. thaliana AtGWD1 (SEX1), AtGWD3 (PWD), a putative Oryza sativa PWD, S. tuberosum StGWD1 and the newly identified S. tuberosum StGWD3 amino acid sequences were built using Clustal X 1.83 (Thompson et al. 1997; Jeanmougin et al. 1998). MSA manual editing and analysis was performed using the GeneDoc 2.6.02 (Nicholas et al. 1997) and Jalview 2.3 (Clamp et al. 2004) MSA analysis tools. Identification of the coding region, open reading frames and the translation of the StGWD3 nucleotide sequence was based on the FGENESH gene structure prediction based on the Hidden Markov Model (HMM) constructed for Nicotiana tabacum (Solanaceae) genome (http://linux1.softberry.com/berry.phtml).
The CBM20 SBD from Bacillus stearothermophilus maltogenic α-amylase (Dauter et al. 1999) (PDB:1QHO, 24 % similarity over amino acid residues 553–780) and the CBM25 carbohydrate-binding module from B. halodurans maltohexaohydrolase (maltohexaose producing amylase; Boraston et al. 2006) (PDB:2C3V, 30 % similarity over amino acid residues 9–90) were used as templates to generate StGWD3 and StGWD1 SBD homology models, respectively. B. stearothermophilus maltogenic α-amylase contains only one starch binding module, classified as member of CBM20 family, whereas B. halodurans maltohexaohydrolase contains two starch binding domains, identified as members of CBM25 and CBM26 families, but only CBM25 was used in this study. Template structures for preliminary models were selected based on scores of the Fold and Function Assignment System (FFAS03) (Rychlewski et al. 2000; Jaroszewski et al. 2005). To investigate the structural alignment of three FFAS03 best scoring StGWD3 SBD homologues (i.e. the granular SBD of Aspergillus niger glucoamylase (PDB:1AC0), the oligosaccharide-binding fragment of Hypocrea jecorina glucoamylase (PDB:2VN4) and the oligosaccharide-binding fragment of the B. stearothermophilus maltogenic α-amylase (PDB:1QHO), the provisional model of the potato SBD was constructed, using automated Swiss-Model mode (http://swissmodel.expasy.org/) with multi-template option. As aligned structures were crystallised with carbohydrate ligands, the preliminary superposition of carbohydrate binding sites enabled the comparative study of CBM20 regions (Fig. 3a) and helped make adjustments to initial FFAS03 alignments.
The proposed preliminary models for potato CBM20 and CBM45 were made using the Schrödinger® Prime 4.5 fully integrated package for homology-based protein structure prediction. The alignments were manually edited. After model building, the chosen loops near the starch-binding sites were refined using the loop refinement module of the Prime package (Jacobson et al. 2004).
To compare the binding energies of the four selected phosphorylated and unphosphorylated oligosaccharide ligands, Schrödinger® Glide 4.5 was used, which is an automated procedure for ligand-receptor docking, in standard and extra-precision mode. Grid-based Ligand Docking with Energetics (Glide) searches was performed for favourable interactions between the protein receptor and putative ligand molecules.
After generating several possible ligand orientations, their interactions with the receptor were evaluated. The best candidates were used in the final step of the algorithm, which was energy minimisation of the ligand-receptor complex in the optimized potentials for liquid simulations—all atom (OPLS-AA) force field. Final scoring using the Glide-score function was carried out on energy-minimised conformations. The ligand positions obtained during this step were then ranked using the Glide-score values (kcal/mol). The elements that make up the Glide-score function are specified below:
where vdW, van der Waals interaction energy; Coul, Coulomb interaction energy; Lipo, lipophilic-contact plus phobic-attractive term; HBond, hydrogen-bonding term; Metal, metal-binding term (usually a reward); Rewards, various reward or penalty terms; RotB, penalty for freezing rotatable bonds; and Site, polar interactions in the active site. The coefficients of vdW and Coul are a = 0.063 and b = 0.120 for Standard Precision (SP) Glide 4.5. CvdW = Coul + vdW is the non-bonded interaction energy between the ligand and the receptor. The Emodel is a specific combination of GScore, CvdW and Intern, where Intern = the internal torsional energy of the ligand conformer.
The receptor structure was considered rigid, and the ligand structures were fully flexible. For each ligand, approximately 30 possible conformations were considered, which were generated using the LLMOD (large-scale low-mode sampling) mode of the Schrödinger MacroModel package.
Results
Sequence analysis and homology modelling
The full-length sequence of StGWD3 cDNA contains a 3,609-bp open reading frame (ORF), a 348-bp 3′ UTR and 198-bp 5′ UTR (GenBank: GU045560.1; GI:270269269). StGWD3 encodes a protein of 1,202 amino acids with transit peptide (Supplemental Fig. S1), (GenBank: ACZ66259.1; GI:270269270) with a calculated molecular mass of 132.27 kDa and an isoelectric point at 5.8.
A BLAST analysis of the StGWD3 amino acid sequence revealed the presence of two conserved domains: a C-terminal PPDK_N nucleotide-binding domain (PEP/pyruvate-binding domain; Pfam: PF01326) and an N-terminal CBM20 starch-binding module (Pfam: PF00686). There is a linker region of ~450 amino acids between these two domains, where a [TL]-S–H phosphohistidine domain (COGs: COG 00391) (PEP utilising enzyme, mobile domain) was identified, which is thought to be mobile in all proteins where it is found (Fig. 1).
The sequence of StGWD3 has 58.6 % identity and 75.3 % similarity to AtGWD3 and shows a similar level of homology to the putative GWD3 from Oryza sativa (59.8 % identity and 75.9 % similarity). The StGWD3 sequence has only 29.7 % identity and 48.2 % similarity to the StGWD1 protein. An alignment of CBM20 domains from three GWD3 sequences (Fig. 2a) shows the conservation of all four important residues from two symmetrically placed SBDs identified in CBM20 region (Machovič and Janeček 2006; Christiansen et al. 2009b, Janeček et al. 2011): W118, K146 and W158 from SBS-1 (within the conserved W-E-X-G-X-N motif (amino acids 158–163 of the StGWD3 sequence) and W132 from SBS-2 (Fig. 2a). An alignment of phosphohistidine and PPDK domains of AtGWD3, StGWD3 and Os12g0297500 sequences (Fig. 2b, c) shows very high sequences identity within this part of analysed sequences.
MSA alignments of GWD sequences. For analyses fragments of three GWD3 sequences, AtGWD3, the newly identified StGWD3, the putative O. sativa GWD3 (GenBank: Os12g0297500), and two GWD1 sequences, StGWD1 and AtGWD1 were used. a The CBM20 starch-binding domain; only the alignment of the GWD3-like sequences is shown. b Alignment of the region of the phosphohistidine domain. c Alignment of the nucleotide-binding domain (PPDK) region (colour figure online)
In our study, homology-modelling templates were chosen based on their FFAS03 score. For the S. tuberosum CBM20 domain model, the top three crystal structures from the PDB database were selected: the granular SBD of A. niger glucoamylase complexed with cyclodextrin (PDB:1AC0), the oligosaccharide-binding fragment of H. jecorina glucoamylase complexed with acarbose-derived hexasaccharide (PDB:2VN4) and the oligosaccharide-binding fragment of the B. stearothermophilus maltogenic α-amylase complexed with maltose (Novamyl) (PDB:1QHO). All proteins mentioned above were co-crystallised with oligosaccharide ligands, which was helpful for identifying the conserved residues involved in sugar binding. According to the CLUSTAL × multiple alignments, there is a highly conserved 6-amino-acid motif in all the sequences mentioned above: WE[NS][GD][PS]N (Fig. 3a).
a Alignment of three best scoring sequences from FFAS03 and the StGWD3 CBM region. Hj_2VN4, Hypocrea jecorina glucoamylase—oligosaccharide-binding fragment; Bs_1QHO, Bacillus stearothermophilus maltogenic α-amylase; Novamyl, oligosaccharide-binding fragment; An_1AC0, granular starch-binding domain of Aspergillus niger glucoamylase. b Multi-alignment of six plant sequences containing two repeats of CBM45 domain with characteristic motif of three tryptophans: first within LHWG conserved motif, second in the distance of 10 amino acids and third in the distance of above 80–100 residues. MSA based on fragments of six plant SBDs from: O. sativa hypothetical protein (Os_hp-EEC80673), S. lycopersicon GWD (Sl_GWD-ACG69788), S. tuberosum StGWD1, V. vinifera hypothetical protein (Vv_hp-CAN69906.1), O. sativa α-amylase (Os_aAMY-NP_001044062) and Z. mays hypothethical protein (Zm_hp-CAW95497) (colour figure online)
The analysis of the ligand-receptor complexes revealed that all residues from the above-described motif are located in proximity to oligosaccharide ligands. After analysis of pairwise alignments between the newly obtained amino acid sequence of StGWD3 and sequences from the three homologous structures, the structure of the oligosaccharide binding-fragment of the B. stearothermophilus maltogenic α-amylase (PDB:1QHO) was selected for homology modelling with the Schrödinger® Prime modelling package, because all of its key residues that form the substrate-binding site are conserved and correspond to the appropriate residues of StGWD3 in the preliminary model.
CBM45 domain has been described so far only in higher plant sequences, none of them has been analysed by means of X-ray or NMR methods and there is no appropriate template structure available in PDB database. Identification of the template for modelling purposes had to be based on FFAS03 (Fold and Function Assignment) scores. To model the CBM45 domain from StGWD1, the carbohydrate-binding module (classified as CBM25) from the B. halodurans maltohexaohydrolase (PDB:2C3V) with score −13.10 (above the critical threshold of −9.00) was selected as a best available template. After analysing the CBM45-family alignments, conserved domains and probable key residues were identified (Fig. 3b). In the selected template structure and in bacterial key histidine motif (LHW) two important tryptophan residues were conserved, what gave high prediction scores. Based on these findings a preliminary homology model of CBM45 domain was constructed.
The CBM45 fingerprint is a tryptophan triad, with one residue within the LHWG motif, the second 10 residues to the right and the third at a distance of approximately 100 amino acids (Fig. 3b). In bacterial CBM25 domain, very similar to the higher plant CBM45, the LHW motif is replaced by IHY, but the remaining two tryptophans are conserved. After aligning the StGWD3 and StGWD1 SBDs with the B. halodurans maltohexaohydrolase sequence, the conservation of the LHW histidine and a conserved similarity of residues in the neighbourhood of the ligand-binding site were observed. The constructed model based on the presented sequence alignment is shown in Fig. 4a, b.
Structural models of StGWD3 and StGWD1 molecules. a Left panel Superposition of the modelled structure of the starch-binding domain (SBD) of potato GWD3 and the template structure of the oligosaccharide-binding fragment of the B. stearothermophilus maltogenic α-amylase (PDB:1QHO) crystallised with its maltose ligand. The ligand-binding site residues within the WE[NS][GD][PS]N[HR] motif are depicted in the wire model (orange ribbons and black residues represent the model; grey ribbons and grey residues represent the template). Right panel zoom of the StGWD3 SBD model. b Left panel Superposition of the modelled structure of the SBD domain in GWD1 and the template structure of B. halodurans maltohexaohydrolase SBD (PDB:2C3W) crystallised with its maltotetraose ligand. The ligand-binding site residues are depicted in the wire model (orange ribbons and black residues represent the model; grey ribbons and grey residues represent the template). Right panel zoom of the GWD1 SBD model. The Trp residues contacting the oligosaccharide are depicted in the stick presentation (gold), and the maltotetraose moiety docked to GWD1 SBD is coloured red (colour figure online)
Analysis of StGWD3 expression
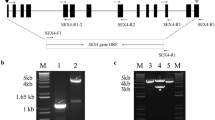
The presence of StGWD3 transcripts (GenBank: GU0455600) was confirmed in the total RNA isolated from both leaves and tubers (Fig. 5). The StGWD3-specific band had a length of 881 bp. Consequently, in further experiments, an attempt was made to analyse the expression of StGWD3 in both hetero- and autotrophic tissues, in tubers at different physiological states and in leaves during the diurnal cycle.
Changes in the levels of StGWD1 and StGWD3 mRNA in leaves in relation to the length of daylight were analysed with PCR and revealed an increased products in real time. This technique allowed a relative assessment of the transcripts levels in reactions with primers specific to fragments of potato EF1-α, StGWD1 and StGWD3 (Fig. 6).
RT-PCR results clearly indicated a change in the amounts of both StGWD1 and StGWD3 transcripts during the day. The expression of StGWD3 during the diurnal cycle increased until 13 h of illumination, when it reached its maximal level. StGWD1 expression reached its maximum after 10 h of illumination, preceding the expression peak of StGWD3. The fluctuations in the expression of StGWD3 during the day were not as prominent as they were for StGWD1. The lowest level of StGWD3 expression was 4 h after turning the light off, and it was more than 2.5 times lower than the maximal expression level. The lowest expression of StGWD1 occurred 4 h after turning the light on, and it was approximately seven times lower than the expression measured 6 h later.
In situ RT-PCR
Considering that the StGWD3 gene is expressed both in potato leaves and tubers, the localisation of its expression was studied in autotrophic leaves and in tubers harvested at an early tuberisation stage or cold-stored. In situ hybridisation procedures did not result in any detectable signal (data not shown); thus, an in situ RT-PCR technique was implemented. The same pair of StGWD3-specific primers was used for in situ RT-PCR as for real-time PCR that gave a 229-bp product (GU0455600). Clear StGWD3 expression signals were obtained on sections of tubers collected at the tuberisation stage and stored in low temperature (Fig. 7a, b, g). The signal was distributed throughout the cytoplasm of all storage parenchyma cells. In leaf samples, no signal was found in young leaves (Fig. 7c), and only a faint signal was observed in fully developed autotrophic leaves (Fig. 7d–f, h).
In situ RT-PCR localisation of StGWD3 transcripts on sections of tubers (a, b, g) and leaves (c–f, h). The expression signal of StGWD3 is visible as a green colouration (a–d, fluorescence microscopy) obtained from FITC fluorescence or as a blue colouration (e and f, light microscopy) obtained from alkaline phosphatase-produced precipitations. No expression signal is present on control sections (g and h, fluorescence microscopy) when primary anti-DIG antibodies were omitted. a and g sections of fresh tuber collected at the tuberisation phase; b section of a cold-stored tuber; c cross-section of young heterotrophic leaf; d, h sections of old autotrophic leaf; e cross-section of autotrophic leaf exposed to light for 3 h; f cross-section of an autotrophic leaf exposed to light for 10 h. Scale bars are 25 μm (colour figure online)
Partial purification of StGWD3 and its identification using MALDI-TOF
We devised a method for purifying StGWD3 from potato tubers, making it possible to obtain a partially purified mixture of proteins that bind specifically to an amylose resin following SDS-PAGE (Fig. 8). One of the protein bands analysed had a molecular mass of approximately 125 ± 3.5 kDa under denaturing conditions.
SDS-PAGE electrophoresis of the partially purified protein fraction obtained during purification of StGWD3. M unstained SDS-PAGE molecular weight marker, H homogenate, As protein fraction after ammonium sulphate fractionation, Ar protein fraction after fractionation on amylose resin. The gel was stained after electrophoresis using Coomassie Blue G-250
The 125-kDa band and the other bands were excised and hydrolysed with trypsin. The molecular masses of the obtained peptides were analysed using MALDI-TOF mass spectrometry. The results were analysed in silico using a procedure similar to that described previously (Ritte et al. 2000; Macewicz et al. 2006). A search of the accessible sequence databases resulted in sequence coverage at 31 %, and 33 out of 68 searched peptide masses matched StGWD3. This protein was used as an antigen for the production of polyclonal antibodies (anti-StGWD3) made by Eurogentec (Belgium).
Immunolocalisation of StGWD3
Western blot analyses of proteins isolated from tubers in different physiological states with a polyclonal antibody directed against StGWD3 indicated the presence of StGWD3 protein in the fraction of soluble proteins isolated from the tubers stored for 3 weeks at room temperature without light (Fig. 9). In the case of proteins isolated from tubers stored at 4 °C for 8 weeks or at a late stage of sprouting (sprouts length exceeded 10–15 cm), no clear immunochemical signal corresponding to the molecular weight of StGWD3 was obtained. Among the proteins isolated from tubers at the initial stage of sprouting (sprouts length approximately 2 cm), StGWD3 was found in the fraction of soluble proteins. There was no immunocytochemical signal for proteins with a molecular weight corresponding to that of StGWD3 in protein fractions isolated from tubers during their formation, when the synthesis of starch granules dominates over its decay. In proteins isolated from leaves exposed to the light for different time periods, no change in the level of StGWD3 was found (data not shown).
Subcellular localisation
Using a polyclonal anti-StGWD3 antibody, the StGWD3 protein was localised in tubers and leaves. Immunocytochemical microscopy indicated that StGWD3 was present in starch grains formed in amyloplasts in tubers (Fig. 10a, b, f). In autotrophic leaves, StGWD3 was present in chloroplasts of palisade and spongy parenchyma cells (Fig. 10c, d). StGWD3 apparently accumulated on transitory starch grains, as the number of labelled chloroplasts increased when the leaves were exposed to light (Fig. 10c vs. d). Transmission electron microscopy showed also sparse gold grains attached to the cell walls (data not shown). This unspecific labelling can explain the faint green colouration of cell walls observed after detection with fluorochrome-conjugated antibodies (Fig. 10a). No fluorescent labelling or colloidal gold grains were observed in starch grains and cell walls when the primary antibody was omitted from the procedure (Fig. 10b, e).
Immunocytochemical localisation of StGWD3. The presence of the StGWD3 protein is indicated by the green colouration (a–e, fluorescence microscopy) obtained from fluorescence of Alexa Fluor488 and colloidal gold grains (f, transmission electron microscopy, some of gold grains are indicated by arrows). a, b, f sections of cold-stored tubers; c-cross-section of an autotrophic leaf exposed to light for 3 h; d, e cross-sections of an autotrophic leaves exposed to light for 10 h; b, e sections of tuber and leaf, respectively, from control labelling when the anti-StGWD3 antibody was omitted. St starch grain. Scale bars a–e 25 μm; f 1 μm (colour figure online)
Discussion
In this study, we provide evidence for the existence of a second glucan, water dikinase (GWD) homologue in the potato, which we named StGWD3. The protein was partially purified and cloned, and the obtained sequences were analysed using bioinformatics methods. The identification of a GWD enzyme activity in the potato and its official naming as α-glucan, water dikinase occurred in 2002 (Ritte et al. 2002). The existence of a GWD was also confirmed in the model organism A. thaliana L. in mutants with increased starch content in the leaves; hence, the A. thaliana homologue was initially named SEX1 (starch excess) (Yu et al. 2001). Nucleotide sequence analysis was used to find GWD1/SEX1 orthologues in A. thaliana. The existence of two additional homologues of AtGWD1 was discovered by a database search (AtGWD2 and AtGWD3). The presence of corresponding signal sequences indicates that only two of them (AtGWD1 and AtGWD3/PWD) are assumed to be localised to plastids (Baunsgaard et al. 2005; Kötting et al. 2005). Mikkelsen et al. (2005) postulated the presence of a GWD homologue in the potato with activity that does not depend on changes in redox potential. Because GWD1 activity is regulated by changes in cellular redox potential, it was assumed that an undiscovered GWD enzyme exists in the potato genome. The presence of a potato EST sequence in the database (CK251207) that exhibits 57 % identity and 73 % similarity with corresponding fragments of AtGWD3 (Mikkelsen et al. 2005) motivated us to purify and clone a second potato isoform of GWD and to perform a bioinformatics analysis of the obtained sequence.
Molecular mass determination
Several protein bands were obtained by modifying a purification protocol for specific binding to an amylose resin, a starch analogue, combined with SDS-PAGE electrophoresis. Using a MALDI-TOF-based identification method, a 31 % sequence coverage of StGWD3 (GU045560) was obtained for a band with a molecular mass of approximately 125 ± 3.5 kDa based on its SDS-PAGE mobility. This molecular mass was similar to the 131.3 kDa of the AtGWD3 molecular mass calculated from the amino acid sequence. The same calculation for StGWD3 results in a molecular mass of 132.3 kDa. In contrast, StGWD1 and AtGWD1 have molecular masses of approximately 160.0 kDa (Ritte et al. 2000) and 146.3 kDa (Yu et al. 2001), respectively.
Bioinformatics analysis of the obtained sequence
According to Christiansen et al. (2009a), starch degradation-related dikinases contain dedicated SBDs at their N-termini that are responsible for substrate binding. The overall domain structure of phosphorylating GWD homologues is similar and, apart from the SBD, always contains a phosphohistidine domain and nucleotide-binding domain (PPDK) (Fig. 2b, c). According to the sequence-based classification, the SBD belongs to the family of carbohydrate-binding modules (CBM indexed from 1 to 64; http://www.cazy.org/fam/acc_CBM.html), which is defined as a contiguous amino acid sequence within a carbohydrate-active enzyme with a discreet fold having carbohydrate-binding activity. For the first time noticeable similarity of N-terminal domains in some starch-degrading enzymes was indicated by Svensson et al. (1989). Subsequent studies revealed that these modules should be divided into several groups named CBM with appropriate indexing. CBM20 is the most thoroughly studied (Machovič and Janeček 2006; Janeček et al. 2011) and is present mainly in amylolytic enzymes from several glycoside hydrolase families, but also in many regulatory enzymes catalysing phosphorylation or acting on phosphate-containing substrates as phosphoglucan, water dikinase (AtGWD3) (Janecek and Sevcík 1999; Janeček et al. 2011), whereas CBM45 (Glaring et al. 2007) was identified in α-amylases and glucan, water dikinase (AtGWD1). The SBDs are 90–130-residues long and typically retain function after isolation. Their conserved structure contains a distorted β-barrel with 7–8 antiparallel strands arranged in two β-sheets. The CBMs bind maltoheptaoses and maltodextrins (Sorimachi et al. 1997; Giardina et al. 2001), but their main function is to attach to the starch granule. Another proposed role is to unwind the α-glucan helices on the granule surface. The functional characteristics of an isolated plant CBM20 showed a relatively low affinity to the starch granule in comparison with fungal CBM20, but analysis of the binding of fluorescein-labelled AtGWD3-SBD to starch granules revealed that AtGWD3-SBD penetrates the interior of these granules more efficiently and is not only limited to binding to the starch granule surface (Christiansen et al. 2009a). Identification of the CBM20 region in the obtained StGWD3 sequence is an important argument in favour of its functional assignment.
Homology modelling of AtGWD3-SBD was attempted by Christiansen et al. (2009a) based on the crystal structure of the SBD of A. niger glucoamylase (GA-SBD; PDB:1KUL and 1AC0). The main structural differences between AtGWD3-SBD and GA-SBD were found in the flexible loop region of binding site 2, which could be responsible for the lower affinity of AtGWD3-SBD to the starch granule.
The results of SBD homology modelling confirm the assumption of SBD-localised differences between the two potato dikinases. After identifying and analysing structures of the bacterial homologues of StGWD1- and StGWD3-SBDs, we constructed hypothetical models of the SBDs for the two potato glucan, water dikinase sequences. Ligand-site residues for the proposed models of potato CBM20 and CBM45 were superimposed onto appropriate template residues (Fig. 4a, b). To test the models, we docked a set of C-6-phosphorylated and unphosphorylated oligosaccharide ligands onto the modelled structures. This experiment was based on the hypothesis that the sequenced StGWD3 (PWD) homologue from the potato has the physicochemical properties of AtGWD3 and that glucan C-6-phosphorylation is necessary for GWD3 (PWD) substrate-binding affinity, as it was proposed in the model of consecutive action of two plant glucan, water dikinases (Fettke et al. 2009).
The results of docking (Glide scores) are presented in Fig. 11. Before docking, we performed a control Glide run with the B. stearothermophilus maltogenic α-amylase (PDB:1QHO) and the B. halodurans maltohexaohydrolase (PDB:2C3V) template structures as receptors for the maltose and maltotetraose ligands that were originally co-crystallised with the above-mentioned proteins, respectively. The values obtained were close to the scores calculated for the appropriate oligosaccharides from the set of examined ligands. In the CBM20 model of StGWD3, both elongation and phosphorylation of oligosaccharide chains, except maltotriose, improved the docking score. The differences in docking scores between the phosphorylated and unphosphorylated forms for a given oligosaccharide were in the range of 20 %. The same range was observed for maltose and maltotetraose docked to the same SBD receptor. These data confirm that the effects of phosphorylation and chain elongation on substrate binding are comparable. The same set of ligands docked to the carbohydrate-binding module modelled for the StGWD1 amino acid sequence yielded similar results for short and longer oligosaccharides. It seems to confirm that the mode of starch binding in the case of CBM45, as well CBM20 is very similar. According to recent findings of Glaring et al. (2011), CBM45 domain binds to starch and soluble cyclodextrin with about twice lower affinity than classical microbial SBDs, such as CBM20 or CBM21. Such findings support the previous hypothesis that low-affinity SBDs are important for dynamic and reversible interactions in starch metabolism (Blennow and Svensson 2010). This has been confirmed by relation between obtained docking scores for CBM45 and CBM20 models. Glide Scores for StGWD3 CBM20 model are relatively higher than scores obtained for CBM45 model. This relation is kept for both phosphorylated and unphosphorylated ligands.
Glide scores for the 16 ligand-receptor pairs. Results for StGWD3 SBD (a) and StGWD1 SBD (b) docking with the phosphorylated and unphosphorylated forms of maltose, maltotriose, maltotetraose or maltopentaose were obtained using Glide 4.5 software with the OPLS-AA force field. Calculations for ligands longer than five glucose rings could not be made because of the internal limitation of the program for the number of atoms and rotatable bonds for ligand molecules
The docking results indicate that the CBMs from StGWD3 and StGWD1 do not differ in their relative affinities for phosphorylated ligands, because phosphorylation positively influenced binding energy. However, there is no evidence that the C-6 phosphate in the glucosyl ring indirectly influences the StGWD3 SBD engagement. Thus, it is possible that the consecutive action of two glucan dikinases is really based on the mechanism proposed by Fettke et al. (2009). In the first step, GWD1 phosphorylates highly ordered, insoluble starch, and glucan phosphorylation at the C-6 position results in a transition state of the phosphoglucans, which is less ordered but still insoluble. This specific state of the starch granule is an appropriate substrate for GWD3 (PWD) catalysis (C-3 phosphorylation), after which the phosphoglucan finally becomes soluble.
The localisation of StGWD3 in various organs and tissues of potato
In potato, the presence of the StGWD1 gene and protein both in leaves and in tubers was confirmed (Lorberth et al. 1998; Mikkelsen et al. 2005). The spatial expression pattern of the StGWD3 gene was analysed in S. tuberosum organs using the in situ RT-PCR method (Fig. 7). The reliable localisation of StGWD3 transcripts was achieved only after applying an in situ RT-PCR method that is more sensitive than in situ hybridisation. In situ RT-PCR followed by fluorescent detection clearly identified StGWD3 mRNA in tubers, but only enzymatic detection indicated that transcripts of the StGWD3 gene were present in leaf parenchyma cells. It seems that the expression level of StGWD3 in leaves is much lower than in tubers. However, in conjunction with the results of in vitro and real-time RT-PCR using a template of total RNA isolated from leaves, the expression of StGWD3 in these organs could be confirmed (Figs. 5, 6). In vitro, RT-PCR had been used previously for organo-specific localisation of AtGWD3 gene transcripts in A. thaliana leaves, roots, stems and inflorescences (Baunsgaard et al. 2005). It was shown that the expression of the AtGWD3 gene occurs in all organs where the starch is present.
The immunochemical localisation of the StGWD3 protein with the use of polyclonal anti-StGWD3 antibody (Fig. 10) showed that StGWD3 in potato is associated with the storage of starch grains formed in amyloplasts and with transitory starch grains formed in leaf parenchyma chloroplasts. In addition, StGWD3 is present mainly in the fraction of chloroplast proteins that are superficially associated with the starch granules. Western blotting following the SDS-PAGE analysis of the abundance of StGWD3 in tubers in different physiological states showed that StGWD3 was abundant in early sprouting potato tubers and less abundant in other analysed organs, including freshly harvested tubers (Fig. 9). Expression pattern of StGWD3 in leaf extracts demonstrated that the level of the StGWD3 protein is independent from the circadian clock (data not shown). It could be due to the lack of high sensitivity of Western blotting method or relatively low level of expression of StGWD3 in potato leaves, because we could observe clearly differences in mRNA levels of StGWD3 and StGWD1 (Fig. 6). Taking in account diurnal cycle in starch degradation in chloroplast, these results suggest a different role of StGWD3 (PWD) in the starch degradation in heterotrophic and autotrophic tissues of potato.
Changes in StGWD3 expression
The analysis of the expression of the StGWD3 and StGWD1 genes in leaves of S. tuberosum throughout the day/night cycle showed that the lowest mRNA levels were observed at the end of the dark period and in the first half of the light period (Fig. 6). Over the next few hours of light availability, there was a significant increase in the amount of both StGWD1 and StGWD3 transcripts. The maximal level of StGWD1 transcript occurred after exposure to light for 10 h and after 13 h for StGWD3. Therefore, a shift between the maximal levels of transcripts for these genes was observed, consistent with the general trend that the expression of both dikinases grows at the end of the light period. Similar diurnal variations were found for the mRNAs of 13 genes, including those encoding DPE2 (EC 2.4.1.25), PHS2 (EC 2.4.1.1), GWD1 and GWD3 in A. thaliana (Smith et al. 2004; Baunsgaard et al. 2005; Kötting et al. 2005). Most of these genes encode proteins that have a confirmed involvement in starch degradation. According to Grennan (2006), a common regulation of the transcription of a gene group may suggest that they participate in the same metabolic pathway; e.g. in starch degradation. Of note, the final activity of individual proteins may be the result of post-translational modifications, such as proteolytic modifications, changes in redox potential or phosphorylation/dephosphorylation (Smith et al. 2005; Orzechowski 2008; Blennow and Svensson 2010). In Arabidopsis leaves, the phosphatase SEX4 (Hejazi et al. 2010) was detected, which acts in opposition to glucan, water dikinases. Phosphatase removes the phosphate groups introduced by AtGWD1 or AtGWD3 activities, allowing the correct degradation of starch by amylases. The occurrence of these regulatory mechanisms of starch degradation does not exclude the presence of a common regulation at the transcriptional level of gene group responsible for the starch mobilisation in vascular plants. However, the sequences that may be responsible for controlling the expression of genes involved in starch metabolism have not been identified.
The role of glucan, water dikinases
The results of in vitro studies with amyloplasts isolated from potato tubers prove that starch phosphorylation is necessary for both its biosynthesis (Wischmann et al. 1999) and degradation (Yu et al. 2001; Baunsgaard et al. 2005; Kötting et al. 2005). The most thoroughly analysed genes in terms of their presumed function in a cell are StGWD1 from S. tuberosum L. and AtGWD1/SEX1 from A. thaliana L. (Ritte et al. 2002, 2004; Mikkelsen et al. 2005, 2006). In contrast, the AtGWD3 from A. thaliana and its functions have been the subject of only a few experimental studies (Baunsgaard et al. 2005; Kötting et al. 2005; Ritte et al. 2006). Fettke et al. (2009) have suggested that GWD3 (PWD) in Arabidopsis acts downstream of GWD1. NMR analyses (Ritte et al. 2006) demonstrated that GWD3 (PWD) selectively phosphorylates the C-3-position of glucosyl residues, whereas GWD1 always introduces the phosphate group at the C-6 position. It is worth emphasising that the C-3-bound phosphate is rarely found in nature (Damager et al. 2010). The recently combined NMR and molecular modelling approaches (Hansen et al. 2008, 2009) have indicated that the C-3 phosphorylation of maltose moieties strongly influences the conformation equilibrium of the maltosidic linkage when the hydration effects are considered (Hansen et al. 2009; Damager et al. 2010). The main effect is presumably steric, but the configuration of the structural water network is also altered, which results in problems with the accommodation of 3′-O-phosphate groups in the helix and leads to a local disruption effect. Hence, the role of C-3 phosphorylation seems to be clear and confirmed by different approaches and studies, but the significance of C-6 phosphorylation remains unclear. According to Blennow and Engelsen (2010), C-6 phosphate groups can propagate signals by tagging particular positions in the starch granule to be locally melted. GWD1 could be a kind of ‘tracker’ that finds and marks the sites required for local amorphisation, which is executed by its GWD3 (PWD) isoform. It is possible that C-6 phosphorylation can induce hydration and voids between helices and thereby facilitate access to the glucosidic bonds for endo-active hydrolases, whereas a more severe disruption of the helix are caused by GWD3 (PWD) activity that stimulates exo-hydrolases and debranching enzymes.
The analysis of the StGWD3 day/night expression cycle in leaves of S. tuberosum, along with earlier studies on the significance of the phosphorylation in in vitro processes, may prove the hypothesis that the phosphorylated substrate changes its conformation through the activity of GWD1/GWD3 (PWD) and becomes more accessible to amylolytic enzymes, thereby may initialise the process of starch grain decomposition (Edner et al. 2007; Dudkiewicz et al. 2008; Hejazi et al. 2008, 2009; Damager et al. 2010). The initial stage of degradation is important because the native starch grain, with its semi-crystalline structure, is not a good substrate for amylases or starch phosphorylase (Tester et al. 2004). The results of the Western blot analysis performed with extracts of tubers in the early sprouting stage (Fig. 9) and the presence of StGWD3 bound to the surface of starch grains in tubers may confirm the key role of StGWD3 in the decomposition of storage starch. In turn, the lack of enzyme bound to starch grains during the tuberisation phase and the lack of any immunocytochemical signal at the late sprouting stage (Fig. 9) may indicate that StGWD3 is not involved in reserve starch biosynthesis and is less involved in the late stage of reserve release in sprouting tubers. Combining our results and the findings of Weise et al. (2011) we could assume that GWD1 is required for phosphorolytic and hydrolytic pathways of starch breakdown as well as for synthesis of starch in leaves, while GWD3 (PWD) is involved in hydrolytic starch mobilisation in tubers. However, the existence of a post-transcriptional mechanism regulating activity of StGWD3 and enzymes of starch metabolism, that is different from the redox mechanism characteristic of StGWD1, is likely (Smith et al. 2004; Mikkelsen et al. 2005). Such regulation could involve the N-terminal domain of StGWD3. It is known that AtGWD3 and AtGWD1 in leaves of A. thaliana differ in substrate specificity due to the N-terminal domain (Kötting et al. 2005; Ritte et al. 2006). Therefore, it is possible that the same domain also plays a regulatory role.
Author contribution
SO: have made substantial contribution to work conception, experimental design and analysis of data; SO, AG, DS, JS, MF, SF: performed the experimental work; MD: has made substantial contribution to bioinformatics analysis; SO, MD and MS have been involved in revising critically the manuscript for important intellectual content. All the Authors read and approved the final manuscript.
Abbreviations
- CBM20 and CBM45:
-
Carbohydrate binding module type 20 and 45
- GWD1:
-
Glucan, water dikinase
- GWD3:
-
Phosphoglucan, water dikinase (PWD)
- SBD:
-
Starch-binding domain
References
Baunsgaard L, Lütken H, Mikkelsen R, Glaring MA, Pham TT, Blennow A (2005) A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated α-glucans and is involved in starch degradation in Arabidopsis. Plant J 41:595–605
Blennow A, Engelsen SB (2010) Helix-breaking news: fighting crystalline starch energy deposits in the cell. Trends Plant Sci 15:236–240
Blennow A, Svensson B (2010) Dynamic of starch granule biogenesis—the role of redox-regulated enzymes and low-affinity carbohydrate-binding modules. Biocatal Biotransform 28:3–9
Boraston AB, Healey M, Klassen J, Ficko-Blean E, Lammerts van Bueren A, Law V (2006) A structural and functional analysis of α-glucan recognition by family 25 and 26 carbohydrate-binding modules reveals a conserved mode of starch recognition. J Biol Chem 281:587–598
Christiansen C, Hachem MA, Glaring MA, Viksø-Nielsen A, Sigurskjold BW, Svensson B, Blennow A (2009a) A CBM20 low-affinity starch-binding domain from glucan, water dikinase. FEBS Lett 583:1159–1163
Christiansen C, Abou Hachem M, Janeček S, Viksø-Nielsen A, Blennow A, Svensson B (2009b) The carbohydrate-binding module family 20-diversity, structure, and function. FEBS J 276:5006–5029
Clamp M, Cluff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20:426–427
Damager I, Engelsen SB, Blennow A, Møller BL, Motawia MS (2010) First principles insight into the α-glucan structures of starch: their synthesis, conformation, and hydration. Chem Rev 110:2049–2080
Dauter Z, Dauter M, Brzozowski AM, Christensen S, Borchert TV, Beier L, Wilson KS, Davies GJ (1999) X-ray structure of Novamyl, the five-domain “maltogenic” alpha-amylase from Bacillus stearothermophilus: maltose and acarbose complexes at 1.7 Å resolution. Biochemistry 38:8385–8392
de Almeida Engler J, De Groodt R, Van Montagu M, Engler G (2001) In situ hybridization to mRNA of Arabidopsis tissue sections. Methods 23:325–334
Dudkiewicz M, Siminska J, Pawłowski K, Orzechowski S (2008) Bioinformatics analysis of oligosaccharide phosphorylation effect on the stabilization of the β-amylase-ligand complex. J Carb Chem 27:479–495
Edner C, Li J, Albrecht T, Mahlow S, Hejazi M, Hussain H, Kaplan F, Guy C, Smith SM, Steup M, Ritte G (2007) Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial amylases. Plant Physiol 145:17–28
Fettke J, Eckermann N, Kötting O, Ritte G, Steup M (2006) Novel starch-related enzymes and carbohydrates. Cell Mol Biol 152(Suppl):OL883–OL904
Fettke J, Hejazi M, Smirnova J, Höchel E, Stage M, Steup M (2009) Eukaryotic starch degradation: integration of plastidial and cytosolic pathways. J Exp Bot 60:2907–2922
Fudali S, Janakowski S, Sobczak M, Griesser M, Grundler FMW, Golinowski W (2008) Two tomato α-expansins show distinct spatial and temporal expression patterns during development of nematode-induced syncytia. Physiol Plant 132:370–383
Giardina T, Gunning AP, Juge N, Faulds CB, Furniss CS, Svensson B, Morris VJ, Williamson G (2001) Both binding sites of the starch binding domain of Aspergillus niger glucoamylase are essential for inducing a conformational change in amylose. J Mol Biol 313:1149–1159
Glaring MA, Zygadło A, Thorneycroft D, Schulz A, Smith SM, Blennow A, Baunsgaard L (2007) An extra-plastidal α-glucan, water dikinase from Arabidopsis phosphorylates amylopectin in vitro and is not necessary for transient starch degradation. J Exp Bot 58:3949–3960
Glaring MA, Baumann MJ, Hachem MA, Nakai H, Nakai N, Santelia D, Sigurskjold BW, Zeeman SC, Blennow A, Svensson B (2011) Starch binding domains in the CBM45 family—low-affinity domains from glucan, water dikinase and α-amylase involved in plastidial starch metabolism. FEBS J 278:1175–1185
Grennan AK (2006) Regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 142:1343–1345
Hansen PI, Larsen FH, Motawia SM, Blennow A, Spraul M, Dvortsak P, Engelsen SB (2008) Structure and hydration of the amylopectin trisaccharide building blocks-synthesis, NMR, and molecular dynamics. Biopolymers 89:1179–1193
Hansen PI, Spraul M, Dvortsak P, Larsen FH, Blennow A, Motawia MS, Engelsen SB (2009) Starch phosphorylation-maltosidic restrains upon 3′- and 6′-phosphorylation investigated by chemical synthesis, molecular dynamics and NMR spectroscopy. Biopolymers 91:179–193
Hejazi M, Fettke J, Haebel S, Edner C, Paris O, Frohberg C, Steup M, Ritte G (2008) Glucan, water dikinase phosphorylates crystalline maltodextrins and thereby initiates solubilisation. Plant J 55:323–334
Hejazi M, Fettke J, Paris O, Steup M (2009) The two plastidial starch-related dikinases sequentially phosphorylate glucosyl residues at the surface of both the A- and B-type allomorphs of crystallized maltodextrins but the mode of action differs. Plant Physiol 150:962–976
Hejazi M, Fettke J, Kötting O, Zeeman SC, Steup M (2010) The laforin-like dual-specificity phosphatase SEX4 from Arabidopsis hydrolyzes both C6- and C3-phosphate esters introduced by starch-related dikinases and thereby affects phase transition of α-glucans. Plant Physiol 152:711–722
Hejazi M, Steup M, Fettke J (2012) The plastidial glucan, water dikinase (GWD) catalyses multiple phosphotransfer reactions. FEBS J 279:1953–1966
Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE, Friesner RA (2004) A hierarchical approach to all-atom loop prediction. Proteins 55:351–367
Janeček S, Sevcík J (1999) The evolution of starch-binding domain. FEBS Lett 456:119–125
Janeček S, Svensson B, MacGregor EA (2011) Structural and evolutionary aspects of two families of non-catalytic domains present in starch and glycogen binding proteins from microbes, plants and animals. Enzyme Microb Technol 49:429–440
Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A (2005) FFAS03: a server for profile-profile sequence alignments. Nucl Acids Res 33:W284–W288
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–405
Kötting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G (2005) Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol 137:242–252
Kötting O, Kossmann J, Zeeman SC, Lloyd JR (2010) Regulation of starch metabolism: the age of enlightenment? Curr Opin Plant Biol 13:1–9
Laemmli UL (1970) Cleavage of structural proteins during the assembly and of the head of bacteriophage T4. Nature 227:680–685
Lloyd JR, Kossmann J, Ritte G (2005) Leaf starch degradation comes out of the shadows. Trends Plant Sci 10:130–137
Lorberth R, Ritte G, Willmitzer L, Kossmann J (1998) Inhibition of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nat Biotech 16:473–477
Macewicz J, Orzechowski S, Dobrzynska U, Haebel S (2006) Is quantity of protein in barley forms determined by proteins localized in the subaleurone layer? Acta Physiol Plant 28:409–416
Machovic M, Janeček S (2006) Starch-binding domains in the post-genome era. Cell Mol Life Sci 63:2710–2724
Mikkelsen R, Baunsgaard L, Blennow A (2004) Functional characterisation of α-glucan, water dikinase, the starch phosphorylating enzyme. Biochem J 377:525–532
Mikkelsen R, Mutenda KE, Mant A, Schürmann P, Blennow A (2005) α-Glucan, water dikinase (GWD): a plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc Natl Acad Sci USA 102:1785–1790
Mikkelsen R, Suszkiewicz K, Blennow A (2006) A novel type carbohydrate-binding module identified in α-glucan, water dikinases is specific for regulated plastidial starch metabolism. Biochemistry 45:4674–4682
Nicholas KB, Nicholas HB Jr, Deerfield DW (1997) GeneDoc: analysis and visualization of genetic variation. EMBNEW.NEWS 4:14. http://www.psc.edu/biomed/genedoc
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914
Orzechowski S (2008) Starch metabolism in leaves. Acta Biochim Pol 55:435–445
Ritte G, Lorberth R, Steup M (2000) Reversible binding of the starch-related R1 protein to the surface of the transitory starch granules. Plant J 21:387–391
Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M (2002) The starch related R1 protein is an α-glucan, water dikinase. Proc Natl Acad Sci USA 99:7166–7171
Ritte G, Scharf A, Eckermann N, Haebel S, Steup M (2004) Phosphorylation of transitory starch is increased during degradation. Plant Physiol 135:2068–2077
Ritte G, Heydenreich M, Mahlow S, Haebel S, Kötting O, Steup M (2006) Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett 580:4872–4876
Rychlewski L, Jaroszewski L, Li W, Godzik A (2000) Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci 9:232–241
Smith AM (2012) Starch in the Arabidopsis plant. Starch 64:421–434
Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 136:2687–2699
Smith AM, Zeeman SC, Smith SM (2005) Starch degradation. Annu Rev Plant Biol 56:73–98
Sorimachi K, Le Gal-Coeffet MF, Williamson G, Archer DB, Williamson MP (1997) Solution structure of the granular starch binding domain of Aspergillus niger glucoamylase bound to β-cyclodextrin. Structure 5:647–661
Svensson B, Jespersen H, Sierks MR, MacGregor EA (1989) Sequence homology between putative raw-starch binding domains from different starch-degrading enzymes. Biochem J 264:309–311
Tester RF, Karkalas J, Qi X (2004) Starch structure and digestibility enzyme-substrate relationship. World’s Poult Sci J 60:186–195
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25:4876–4882
Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76:4350–4354
Truszkiewicz W, Paszkowski A (2005) Some structural properties of plant serine:glyoxylate aminotransferase. Acta Biochim Pol 52:527–534
Weise SE, van Wijk KJ, Sharkey TD (2011) The role of transitory starch in C3, CAM, and C4 metabolism and opportunities for engineering leaf starch accumulation. J Exp Bot 62:3109–3118
Wischmann B, Nielsen TH, Møller BL (1999) In vitro biosynthesis of phosphorylated starch in intact potato amyloplasts. Plant Physiol 119:455–462
Yu TS, Kofler H, Häusler RE, Hille D, Flügge UI, Zeeman SC, Smith AM, Kossmann J, Lloyd J, Ritte G, Steup M, Lue WL, Chen J, Weber A (2001) The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell 13:1907–1918
Acknowledgments
Work was partly funded by the Polish Ministry of Science and Higher Education (Grant No. N302061134). We thank Malgorzata Wasilewska-Gomulka for sectioning for transmission electron microscopy.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. A. Kleczkowski.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. S1 Amino-acid sequence of StGWD3 protein obtained using FGENESH algorithm (Softberry) based on StGWD3 nucleotide sequence. Different functional domains of the GWD3 sequence are depicted in colours: orange- transit peptide, blue—predicted CBM20 domain, green-phosphohistidine domain, red- PPDK domain.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Orzechowski, S., Grabowska, A., Sitnicka, D. et al. Analysis of the expression, subcellular and tissue localisation of phosphoglucan, water dikinase (PWD/GWD3) in Solanum tuberosum L.: a bioinformatics approach for the comparative analysis of two α-glucan, water dikinases (GWDs) from Solanum tuberosum L.. Acta Physiol Plant 35, 483–500 (2013). https://doi.org/10.1007/s11738-012-1091-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11738-012-1091-y