Abstract
Drought is the most significant abiotic stress in agriculture; thus, this area of studies seems to be one of the most important challenges in plant biology. Data about gene expression under drought are crucial to study drought response mechanisms and to select the genes for a transgenic approach. Quantitative RT-PCR is a powerful method for gene expression analysis; however, obtaining proper data normalization requires internal reference genes with stable level of expression. In the present paper ten potential reference genes were examined in two developmental stages of barley for their expression stability during leaf growth and increasing drought level. The results indicated that leaf growth per se affects the expression of studied genes to the similar extent as the drought and showed that different genes were most stably expressed in the seedling and the heading stage. As a result, different sets of reference genes were selected for different applications. For instance, ADP-ribozylation factor 1 and ubiquitin encoding genes were most suitable to study drought-induced changes in gene expression at the seedling stage, whereas actin and GAPDH genes were useful during heading, and ADP-ribozylation factor 1 and HSP90 allowed for the comparison between these two stages. Our data proved the necessity for validation of commonly used reference genes. The results indicate that expression of ADP seems to be the least affected by all the factors studied in the present experiment. However, when the effect of only one factor among those investigated in this work will be studied, different genes should be considered to be used as the references due to the higher stability of their expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Barley (Hordeum vulgare) is one of the most important cereal crops grown in many areas where it is often subjected to drought stress which significantly affects yielding (Ceccarelli et al. 2007). Despite long-term studies it is still a real challenge to improve crop productivity under water-limiting conditions. The reason is that in contrast to plant resistance to biotic stress the response to abiotic stress is a complex and multigene-controlled mechanism (Vinocur and Altman 2005). Moreover, a strong interaction is observed between plant response to drought and environmental as well as developmental factors. Thus, to improve drought tolerance a better understanding of the genetic bases and the mechanism of drought response is required (Cattivelli et al. 2008). To address this issue studies of the drought-induced changes in the expression of genes involved in signaling and regulatory pathways or genes encoding proteins related to stress tolerance are performed (Vinocur and Altman 2005; Guo et al. 2009; Rapacz et al. 2010). The expression of these genes may be analyzed at the transcription stage using different techniques. However, real-time RT PCR is the most recommended method for the quantitation of mRNA transcripts (Gachon et al. 2004). In both absolute and relative real-time PCR quantification internal standards are needed as the most commonly used normalization method, which enable to exclude the variability of cDNA quantity deriving from sampling storage mRNA extraction and RT protocols (Pfaffl 2004; Huggett et al. 2005). Internal standards or reference genes should reveal a constant level of the expression in a cell or tissue studied and the expression should not change under experimental treatment (Gilsbach et al. 2006). Genes involved in basic cellular functions known as house-keeping genes have been used as reference genes; however, many reports have shown a large variation in their expression (Singh and Green 1993; Radonić et al. 2004; Dheda et al. 2004). As a result, new potential internal standards are prospected. For example, the experimental verification of SAM, HSP90, EF1 and ADP as reference genes was suggested in barley on the basis of in silico analysis (Faccioli et al. 2007). It was widely reported that the expression stability of plant reference genes may be differentially affected by environmental stresses (Nicot et al. 2005; Paolacci et al. 2009; Schmidt and Delaney 2010), the type of tissue (Schmidt and Delaney 2010; Jian et al. 2008; Mallona et al. 2010; Wan et al. 2010), plant species or even genotype used in the studies (Mallona et al. 2010). Thus the selection of proper reference gene(s) for real-time RT-PCR data normalization seems to be one of the most important steps in each gene expression analysis and the stability of potential reference genes should be checked for each environmental factor tissue type and plant species (Guenin et al. 2009). The identification of the most stable gene in a set of assumed internal standards is possible using different softwares based on different statistical protocols such as geNorm (Vandesompele et al. 2002, 2009) developed into qBase Plus (Hellemans et al. 2007) NormFinder (Andersen et al. 2004) or BestKeeper (Pfaffl et al. 2004). As distinct approaches were used in different methods the final identification may vary between them; therefore, the use of different programs is recommended (Schmidt and Delaney 2010; Mallona et al. 2010; Andersen et al. 2004).
To facilitate the further studies on the genetic basis of drought tolerance in barley the real-time RT-PCR based verification of potential reference genes were performed in barley leaves subjected to drought. The studies included two developmental stages in which barley is most affected by drought under Middle-European conditions. To the best of our knowledge we are the first to examine the effects of leaf age by means of the dynamic studies of gene expression during increased drought level and parallel control.
Materials and methods
Plant material and experimental design
The experiments were performed on six-row spring barley (Hordeum vulgare L.) cultivar Morex. Seeds were obtained from the genetic bank of the Institute of Plant Genetics Polish Academy of Sciences in Poznan.
The studies were performed in two independent twice-repeated experiments. Plants were sown in 6,700 cm3 (37 cm long, 14 cm wide and 13 cm high) pots filled with a mixture of clay, peat and sand (3:2:1v/v/v) with nine plants each (three pots for each experimental variant) and grown in a fully controlled growth room at the temperature 25/17°C (day/night)a 16-h photoperiod Photosynthetic Active Radiation (PAR) of 450 μmol (quanta) m−2 s−1 (provided by high-pressure sodium lamps 400 W; Philips SON-T AGRO, Brussels, Belgium) and 40% air humidity. Pots were weighted and filled with the same amount of soil. The soil water content was kept at 70% of the maximum water capacity (MWC) by adding an appropriate amount of water on a daily basis. Once a week plants were fertilized with Florovit multipurpose liquid fertilizer (Inco-Veritas, Gora Kalwaria, Poland). Plants were grown for approximately 3 weeks to reach the phase of the third leaf (seedling stage Experiment 1) or for approximately 6 weeks to reach the beginning of heading (Experiment 2). After reaching the appropriate developmental stage, the watering was stopped in one part of pots (drought treatment), whereas in the control plants the watering regime remained unchanged. In drought-treated plants, water content in the soil decreased gradually reaching 60% of the maximum water capacity (MWC) in the first, 49% in the third, 39% in the fifth and finally 31% (soil water potential: −2 MPa) on the seventh day of drought treatment. The same watering level was maintained until the tenth day. The homogeneity of soil water content in pots was controlled in different points of the pot using the Hydro Sense Soil Water Content Measurement System (Campbell Scientific, Inc., Australia Pty. Ltd.). Additionally, the soil water potential was measured with a HR-33T dew point microvoltmeter supplied with a C-52 thermocouple psychrometer chamber (both by Wescor Inc., Logan, UT, USA).
Samples (0.03–0.05 g from the middle part of the youngest but fully developed leaf) were collected on the first, third, fifth, seventh and tenth day of the drought treatment, always in the fourth hour of the light period both in drought-treated and control plants. There were three biological samples (one from each pot) collected simultaneously in each experiment and in each experimental variant (developmental stage drought treatment/control day of sampling). Samples were frozen in liquid nitrogen immediately after collection and stored in −80°C until use.
Genes
Ten candidate genes were selected as potential housekeeping genes (Radonić et al. 2004; Faccioli et al. 2007). Barley sequences were found in GenBank: actin (ACT)-GenBank ID: AY145451; S-adenosylmethionine decarboxylase (SAM)-GenBank ID: BE413438.1; glycolitic glyceraldehyde-3-phosphate dehydogenase (GAPDH)-GenBank ID: X60343.1; cytosolic heat shock protein 90 (HSP90)-GenBank ID: AY325266.1; alpha tubulin (L-TUB)-U40042.1; beta tubulin (B-TUB)-GenBank ID: AM502854.1; elongation factor 1-α (EF1)-GenBank ID: AJ472912; ubiquitin (UBI)-GenBank ID: M60175.1; ribosomal protein L2 (RPL2)-GenBank ID: X78185.1; ADP-ribosylation factor 1-like protein (ADP)-GenBank ID: AJ508228.2.
Primers were designed in Primer Express Software v. 2.0 (Applied Biosystems, Foster City, CA, USA). Primers were optimized and supplied by Applied Biosystems (Table 1).
Real-time PCR analysis of genes expression
Total RNA was isolated using an RNeasy Plant Mini Kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s instruction. The removal of genomic DNA contamination and first-strand cDNA synthesis were performed using QuantiTect Reverse Transcription Kit (Qiagen) in accordance with the manufacturer’s instructions. The final concentration and quality of cDNA were determined spectrophotometrically (Nanodrop 2000c, Thermo Scientific, Wilmington, DE).
PCR reactions were 25 μl in volume and contained 12.5 μl Power SYBR Green PCR Master Mix (Applied Biosystems by Life Technologies, Carlsbad, CA, USA) 2.5 μl cDNA solution (equivalent to ~10 ng template) 2.5 μl of each primer (final concentration 900 nM) and 5 μl of nuclease-free water (Qiagen). PCR was performed using a 7500 Real-Time PCR System (Applied Biosystems) in 96-well plates with the following cycling parameters: 10 min at 90°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C (Table 1). PCR reactions containing cDNA or ‘no template’ control (NTC; ~15 ng DNase-treated RNA only) were run in parallel for each template and primer combination. The specificity of PCR products was verified by dissociation curve analysis (60–95°C). Each PCR reaction was repeated three times; therefore, the final number of repetition for each experimental variant was three technical instrumental replicates × two repeated experiments × two biological samples (12). Additionally, the third biological sample was also analyzed when C t values varied/differed more than by 10% (approx. 2.5 depending on depending on an experimental object/gene) between the two biological samples. Either extreme result was discarded or all the three results were taken into consideration. Thus the maximum number of total replicates was 18. In order to determine the arbitrary amount of cDNA template in each sample which is required as an initial data for most methods used in the determination of expression stability standard curves were prepared using five different dilutions of the template (Larionov et al. 2005).
The stability of housekeeping gene expression was analyzed using qBasePLUS-geNormPLUS and Quality Control modules (Hellemans et al. 2007) NormFinder (Andersen et al. 2004) and BestKeeper (Pfaffl 2004).
Results
Leaf growth and developmental stage
The analysis of standard curves (r 2 values were higher than 0.99 with the exception of B-TUB: 0.966) showed that amplification efficiency ranged from 1.89 to 2.00 with the exception of B-TUB where it was estimated for 2.71 (data not shown). This gene displayed the presence of non-specific amplification product during melting curve analysis; thus it was not taken into account in further analysis (data not shown).
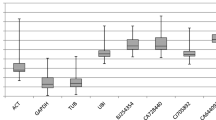
The expression of genes studied in the present experiment was clearly affected by the leaf growth as well as by developmental stage of plants (Table 2; Fig. 1). According to CV analysis the most stable gene at the third leaf stage was UBI. ACT and GAPDH were recommended to be two references in one experiment by geNormPLUS. Other methods, however, classified GAPDH as a rather unstable gene. NormFinder pointed at ADP and UBI as clearly more stable than the other genes whereas according to BestKeeper RPL2 was the best referencewhile ADP had a relatively small SD value as well. When the values of M (Fig. 1) CVSV and SD (Table 2) are compared for the most stable and unstable genes, it is clearly visible that during the heading stage the stability of the expression was generally lower than in the seedling stage. In this case geNormPLUS recommended using three (UBI, GAPDH, ADP) instead of two genes together as references in one experiment. At the heading stage UBI and GAPDH were top ranked by the vast majority of methods (with an exception of UBI in BestKeeper). BestKeeper ranked ACT as the best reference but SD value exceeded in this case that for the most unstable (SAM) at the seedling stage. Moreover, all the values were higher than one which might indicate their inconsistency (Pfaffl et al. 2004). HSP90 appeared to be the most unstable gene at this stage. GeNormPLUS recommended using as many as four genes (ADP, UBI, GAPDH and EF1) for the normalization in search of reference genes for the comparison between two developmental stages. The M values highly exceeded those of a single stage analysis (Fig. 1; Table 2). ADP and UBI showed relatively high stability of NormFinder’s SV. On the other hand, the values of CV and SD were high for each gene. RPL2 which was ranked as the most stable according to BestKeeper was the most unstable in other methods while SD value for RPL2 was higher than one.
The gene expression stability measure (M) calculated with geNormPLUS for growing barley leaves in two developmental stages. Samples were collected on the first, third, fifth, seventh and tenth day of the experiment from the youngest but expanded leaf. During this period no symptoms of aging were observed. M values for potential references useful for the comparison between different developmental stages were also shown
Drought treatment
Studies held in gradually increased drought led to obtaining different results of gene expression stability for the two developmental stages (Fig. 2; Table 3). The most constantly expressed gene at the third-leaf stage was UBI. It was top ranked in qBase CV and NormFinder analysis. In both methods ADP was selected as the second stable gene. UBI and ADP were ranked as the third and the fourth most stable genes in geNormPLUS analysis with M values only slightly higher when compared with the most stable genes. However, geNormPLUS analysis suggested the use of two different genes (RPL2 and EF1) for gene normalization in a single assay. EF1 was also ranked as the second (after HSP90) most stable gene by BestKeeper. In the same method SD value for UBI, top ranked in other methods, was almost two times higher than that for HSP90. As shown for the control the overall variation of the expression in the heading stage was higher than that observed at the third-leaf stage. GeNormPLUS suggested the use of as many as five references (EF1, L-TUB, UBI, HSP90 and ADP). According to qBase and NormFinder L-TUB which was bottom ranked in plants stressed at the seedling stage was the most stable gene when drought treatment was performed during heading. Among the genes most stable in drought-treated seedlings, ADP, but not UBI, was also top ranked in plants stressed at the heading. RPL2 (the only gene with SD below 1) and HSP90 (as in seedling) were top ranked by BestKeeper while RPL2 was bottom ranked by other methods. The most appropriate gene for the comparison of the gene expression level in barley subjected to drought in both seedling and heading stages seems to be ADP. SAM proved to be the least stable in every method. According to geNormPLUS target genes expression analysis in both stages requires four references (EF1, UBI, HSP90 and ADP).
The gene expression stability measure (M) calculated with geNormPLUS for barley leaves drought-treated in two developmental stages. The mean stabilities for both stages were also shown. Samples were collected from the youngest but expanded leaf on the first (60% MWC) third (49% MWC) fifth (39% MWC) seventh (31% MWC) and the tenth (31% MWC) day of the drought treatment
The overall effect of drought on the expression stability of potential reference genes was similar to those observed for leaf growth in the control (Figs. 1, 2; Tables 2, 3). Coefficient of variation values were always lower in drought-treated than in control plants. M values in drought-treated plants were lower than in control for bottom ranked genes but higher for the top ranked. According to BestKeeper the expression of reference genes was generally more stable in drought-treated than control plants at the heading stage, but less stable at the seedling stage. On the other hand SV values of NormFinder were always lower for the control than drought-treated plants.
In the studies of the drought-induced gene expression its level is often calculated as the fold changes relative to the control (well watered plants) (Guo et al. 2009; Atienza et al. 2004; Abebe et al. 2010); thus the comparison of potential reference gene expression stability was also performed for both control and drought-treated plants either separately for each day of the treatment or for all the days taken together into account (general analysis). The rankings obtained for particular days of treatment were slightly different and general analysis pointed out huge variation in every gene expression (data not shown). Finally, we decided to focus on the tenth day of drought treatment when the changes in the expression of HVA1 and SRG6 (drought regulated genes) observed between the control and drought-treated plants as well as differences between their relative expression levels in barleys of different drought tolerance were the highest irrespective of the developmental stage (data in preparation). Here a clear turgor loss was visible in drought-treated plants; however, all the plants survived after rewatering. In most cases the observed variation in the expression level of genes studied here was the lowest when compared with the variation observed either during leaf growth in the control or during drought treatment. All the measures of gene expression stability (MCVSV and SD) were in this case the lowest for top ranked genes (Figs. 1, 2, 3; Tables 2, 3, 4). At the third-leaf stage UBI and ADP were characterized with the most stable expression between the control and drought-stressed plants at the tenth day of the experiment. During heading ACT was the least variable gene (with the exception of BestKeeper ranking). Also geNormPLUS recommended ACT together with GAPDH (which was however the most unstable in the seedling stage) for normalization of gene expression at the heading stage. SAM was characterized with the lowest stability at this stage. For the experiments in which the drought-induced fold changes in gene expression should be compared between seedling and heading stage on the tenth day of drought treatment the use of ADP and HSP90 should be recommended according to geNormPLUS although ADP undoubtedly displayed the highest stability of expression level.
The gene expression stability measure (M) calculated with geNormPLUS for drought-induced changes in barley leaves. Data was calculated for two developmental stages separately as well as the stability was checked for both stages together to select potential references suitable for the comparison between drought treated plants at different stages. Data from the tenth day of both control and drought-treated plants were used in the calculations
Discussion
Methods for the identification of stable reference genes are based on distinct models and often produce different results from the same data set (Paolacci et al. 2009; Schmidt and Delaney 2010; Mallona et al. 2010). On the other hand in the present paper rankings created by CV analysis geNormPLUS (M) and NormFinder (SV) were very similar, while BestKeeper’s ranking was almost always distinct. Similar differences between BestKeeper and other methods were previously reported (Mallona et al. 2010). The most characteristic feature for BestKeeper’s rankings created in the present paper was that RPL2 was in vast majority the most stable in the expression irrespective of the data set analyzed, whereas other methods classified it with some exceptions as one of the most unstable genes. We have found that the mean C t value for RPL2 was the smallest among all the genes (20.95 in comparison to 23.76 for all the other genes). According to the authors of this method slightly expressed genes with C t values of about 30–35 certainly show different (higher) variance compared with highly expressed genes with C ts around 15 or even less (Pfaffl et al. 2004). The author suggested new versions of the software in which Spearman and Kendall Tau correlation should be used for ranking purposes. Unfortunately those versions are not currently available. Additionally, our study showed the possibility of some problems occurring when the differences in C t values are smaller than those indicated by the authors. Different input data required for different methods were also suggested as a major source of the variation between rankings created in different methods (Jian et al. 2008; Mallona et al. 2010). BestKeeper and NormFinder examine C t values whereas qBasePLUS evaluates calculated template amount.
In consequence, the differences in PCR efficiency may affect variation in stability rankings between programs requiring different input data. In our study rankings created by qBasePLUS and NormFinder were very similar. This might be explained by the fact that all amplification efficiencies observed here were very similar and close to two (100%).
The relative expression instability of the potential reference genes observed in our experiments during 10 days of barley leaf growth was comparable or even higher to those observed during drought treatment. This was rather surprising especially because in the beginning of the experiment leaves were expanded and at the end no visual symptoms of aging were observed. The huge impact of leaf growth on the expression of genes studied also prompted some further consequences. The most stable expression of many genes was observed when control and drought-treated plants were compared together at the same time point of the experiment. In this case an important source of the variation leaf growth was excluded. On the other hand, this instability impeded the proper selection of internal standards for dynamic studies of fold (relative to the control) changes in accumulation of transcripts during drought treatment. In this case an additional variation source was added—different leaf growth rates in drying and well-watered plants. Changes in gene expression during the middle period of leaf growth were not studied before while such changes were widely studied during early stages of leaf development and during senescence (Atienza et al. 2004; Druka et al. 2006; Gregersen and Holm 2007). The stability of potential reference gene expression was compared in leaf buds and aging leaves of Petunia hybrida (Mallona et al. 2010). The observed variation in gene expression was certainly higher than in the case of our results (for instance geNorm M values for the most stable gene was 0.77 whereas in our case 0.36) and EF1 was most frequently selected as the most stable gene by those authors (depending on statistics and genotype). In our study EF1 was also found in the group of less variable genes especially during heading and for the whole control (not subjected to drought) dataset thus in the cases when higher overall variation was observed. However, on the basis of our experiment UBI and ADP should be recommended as the internal standards for real-time RT-PCR studies of gene expression in developing leaves of barley. Interestingly, translation products of these both genes are involved in protein processing which seems to be a process rather stable during leaf growth (Liu et al. 2008). On the other hand, three most unstable genes L-TUBRPL2 and HSP90 play rather distinct biological functions (Radchuk et al. 2008; Marty and Meyer 1992; Wang et al. 2004).
The larger variation in the expression levels of nearly all of the studied genes was observed during heading rather than in the third-leaf stage. Two possible explanations may be suggested. (1) The first is a very simple one. As shown earlier the expression of our potential references changed during leaf growth; thus leaf developmental age affects the level of its expression. When sampling is performed at the third-leaf stage we can be confident that those leaves were developed approximately at the same time. At the heading stage due to natural variation in plant growth and development rates the leaf developmental age may be much different in different plants in the beginning of the experiment; thus the level of overall variation observed would be higher. (2) The second factor that may affect larger variation in the expression of genes studied here is the development of generative organs during heading which affects the expression of a large set of genes (Druka et al. 2006). The transcript level of ACT the gene which is very often used for gene expression normalization in plants and reported as stably expressed in vegetative tissues of Arabidopsis (An et al. 1996) was quite stable in seedling although it can be recommended as internal standard neither at the heading stage nor for comparisons between different developmental stages. On the other hand, GAPDH was the only gene which increased their stability during heading when compared with the seedling stage where it was indicated as stable on the basis of M statistics only. It is likely that the activity of glycolitic GAPDH which integrated many metabolic pathways in the cells gets more stable when sink-source relations and leaf metabolism becomes more stable after flowering induction and heading (Wardlaw 1990).
The expression of ADP was not only very stable at the control but also hardly affected by drought treatment both when the transcript level was studied in subsequent days of the treatment as well as when the level of the expression was compared between drought-treated and control plants. Additionally its stability was not affected by development stage. Our results confirmed the suggestion based on in silico studies that ADP may be a good reference gene in barley (Faccioli et al. 2007). UBI the expression level of which was quite stable under control conditions still remained the most stable gene when drought was applied in the seedling stage; however, it was characterized with lower stability during heading. The same was observed when the expression level of this gene between drought-treated and control plants in the last day of the experiment was compared. The results of our experiments showed that transcriptional regulation of UBI seems not to be the element of drought response in general although it may be connected with some interactions between drought signal and leaf development. As suggested earlier the potential role of ubiquitination in abiotic stress would be the regulation of the lifetime of rather signaling molecules than proteins directly connected to stress tolerance (Xiong and Zhu 2001).
In the present study ACT was shown to have very good stability between control and drought-treated plants at the one time-point; however, only during heading. Thus, the stability of ACT expression in drought seems to be dependent on developmental stage. The effects of abiotic stresses on ACT expression were also previously reported (Nicot et al. 2005; Paolacci et al. 2009). On the other hand it was the evidence provided for the stability of ACT in tobacco exposed to several abiotic stresses including drought treatment (Schmidt and Delaney 2010). Furthermore, in the previous studies of our team ACT was used as the reference gene in the studies of drought response at the seedling stage in barley (Rapacz et al. 2010). According to the present results ACT seems not to be the best internal standard for the studies of drought-induced fold increase in gene expression at that developmental stage. In the paper mentioned earlier the final results were calculated as relative to the reference genotype due to the lack of HVA1 expression observed in the control plants (Rapacz et al. 2010). Additionally, that study was performed on the seventh day of drought treatment when the water deficit in leaves was not as substantial as in the current study and the expression of ACT was more stable between drought-treated and control plants (data not shown).
α-Tubuline is one of the major elements of the microtubules which are involved in several cellular processes such as cell shape specification cell transport cell motility cell division and expansion (Radchuk et al. 2008); thus any changes in its expression triggered by factors affecting growth rate (as a drought) should be expected. The previous report demonstrated variability of L-TUB expression under drought in cucumber (Wan et al. 2010). In the present study the effect of drought on the expression level of L-TUB clearly depends on developmental stage and as in the case of ACT higher stability was observed at the heading stage.
SAM, HSP90 and EF1 were suggested as potential internal standards for qRT-PCR studies in barley (Faccioli et al. 2007). In our experiments SAM was the least stable in the vast majority of calculations involving samples collected from drought-treated plants. It can be expected since the involvement of S-adenosylmethionine decarboxylase in polyamines synthesis and polyamine metabolism changes significantly in various abiotic stresses (Alcazár et al. 2006). Usually HSP90 and EF1 were ranked under the best references; however, the variation observed for their expression was lower than that of SAM. These results are contrary to the results of other studies in which the stability of EF1 expression under drought and other abiotic stresses was proved (Faccioli et al. 2007; Nicot et al. 2005; Wan et al. 2010). In fact this may be surprising due to large changes in protein metabolism observed under different abiotic stress conditions (Kosmala et al. 2009; Rasoulnia et al. 2010). One of the stress-induced proteins is HSP90. It was shown that the expression of encoding gene in Arabidopsis is regulated by heat cold salt stress heavy metals and light–dark transitions where management of protein folding seems to be very important (Krishna and Gloor 2001). In the present study the variation in HSP90 expression is, however, more significant during leaf growth (in the control) than under drought and HSP90 was even selected as one of the most stable reference for fold change studies relative to the control. It is possible that protein repair processes are not indispensable during drought. The changes observed in HSP90 transcript levels during leaf growth might be connected with the different functions of encoded protein (signal-transduction networks cell-cycle control protein degradation and protein trafficking) (Wang et al. 2004).
GAPDH was characterized with stable expression level in barley as well and it was used as the stable reference in the study of drought-affected gene expression (Faccioli et al. 2007; Abebe et al. 2010). In the present study, the stability of GAPDH observed in drought was strongly connected with the developmental stage and in seedling this gene was definitely the least stable in its expression when compared with the control level. However, in the study cited above the drought treatment lasted just for 4 days (Abebe et al. 2010). Therefore, the differences in the expression levels between drought-treated and control plants may be insignificant at that time. It seems to be possible because on the basis of our results the expression level of GAPDH seems to be more affected by leaf growth than drought per se.
RPL2 the last of genes studied was previously indicated as the best reference gene in potato plants subjected to abiotic stress treatment (Nicot et al. 2005). This result was not confirmed in our experiments.
Conclusion
The results indicated that leaf growth per se affects the expression of studied genes to the similar extent as the drought. Furthermore the different genes were the most stably expressed at the seedling and heading stage. As a result different sets of reference genes were selected for different applications. The expression level of ADP seems to be the least affected by all the factors studied in the present experiment; however, different genes were proved to be more stable in some instances.
Author contribution
M. Rapacz was responsible for experimental design and results interpretation coordinated data analysis and wrote the paper. A. Stępień and K. Skorupa ran the experiment, performed qPCR studies and data analysis and were also involved in paper preparing.
Abbreviations
- ACT :
-
Actin
- ADP :
-
ADP-ribosylation factor 1
- B-TUB :
-
β-Tubulin
- C t :
-
(Cycle threshold) the mean number of cycles required for the fluorescent signal to cross the threshold
- CV:
-
Coefficient of variation (calculated in qBase)
- EF1 :
-
Elongation factor 1-α
- GAPDH :
-
Glycolitic glyceraldehyde-3-phosphate dehydogenase
- HSP90 :
-
Heat shock protein 90
- HVA1 :
-
H. vulgare Late Embriogenesis Abundant (LEA) 3 family gene 1
- L-TUB :
-
α-Tubulin
- M:
-
Gene expression stability measure of geNormPLUS
- MWC:
-
Maximum water capacity
- RPL2 :
-
Ribosomal protein L2
- SAM :
-
S-Adenosylmethionine decarboxylase
- SD:
-
BestKeeper standard deviation
- SRG6 :
-
Stress-responsive gene 6
- SV:
-
NormFinder stability value
- UBI :
-
Ubiquitin
References
Abebe T, Malmaiee K, Berg V, Wise RP (2010) Drought response in the spikes of barley: gene expression in the lemma, palea, awn and seed. Funct Integr Genomics 10:191–205
Alcazár R, Marco F, Cuevas JC, Patron M, Ferrando A, Carrasco P, Tiburcio A (2006) Involvement of polyamines in plant response to abiotic stress. Biotechnol Lett 28:1867–1876
An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10:107–121
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Atienza SG, Faccioli P, Perrotta G, Dalfino G, Zschiesche W, Humbeck K, Stanca AM, Cattivelli L (2004) Large scale analysis of transcripts abundance in barley subjected to several single and combined abiotic stress conditions. Plant Sci 167:1359–1365
Cattivelli L, Rizza F, Badeck FW, Mazzucotelli E, Mastrangelo AM, Francia E, Marè C, Tondelli A, Stanca AM (2008) Drought tolerance improvement in crop plants: an intergrated view from breeding to genomics. Field Crop Res 105:1–14
Ceccarelli S, Grando S, Baum M (2007) Participatory plant breeding in water-limited environments. Exp Agri 43:1–25
Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37:112–119
Druka A, Muehlbauer G, Druka I, Caldo R, Baumann U, Rostoks N, Schreiber A, Wise R, Close T, Kleinhofs A, Graner A, Schulman A, Langridge P, Sato K, Hayes P, McNicol J, Marshall D, Waugh R (2006) An atlas of gene expression from seed to seed through barley development. Funct Integr Genomics 6:202–211
Faccioli P, Ciceri GP, Provero P, Stanca AM, Morcia C, Terzi V (2007) A combined strategy of “in silico” transcriptome analysis and web search engine optimization allows an agile identification of reference genes suitable for normalization in gene expression studies. Plant Mol Biol 63:679–688
Gachon C, Mingam A, Charrier B (2004) Real-time PCR: what relevance to plant studies. J Exp Bot 55:1445–1454
Gilsbach R, Kouta M, Bönisch H, Brüss M (2006) Comparison of in vitro and in vivo reference genes for internal standardization of real-time PCR data. Biotechniques 40:173–177
Gregersen PL, Holm PB (2007) Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biot J 5:192–206
Guenin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L (2009) Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot 60:487–493
Guo P, Baum M, Grando S, Ceccarelli S, Bai G, Li R, von Korff M, Varshney RK, Graner A, Valkoun J (2009) Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J Exp Bot 60:3531–3544
Hellemans J, Mortier G, Paepe AD, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time PCR data. Gen Biol 8:R19
Huggett I, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalization; strategies and considerations. Genes Immun 6:279–284
Jian B, Liu B, Bi Y, Hou W, Wu C, Han T (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9:59
Kosmala A, Bocian A, Rapacz M, Jurczyk B, Zwierzykowski Z (2009) Identification of leaf proteins differentially accumulated during cold acclimation between Festuca pratensis plants with distinct levels of frost tolerance. J Exp Bot 60:3595–3609
Krishna P, Gloor G (2001) The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 6:238–246
Larionov A, Krause A, Miller W (2005) A standard curve based method for relative real time PCR data processing. BMC Bioinf 6:62–77
Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20:292–306
Mallona I, Lischewski S, Weiss J, Hause B, Egea-Cortines M (2010) Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol 10:4
Marty I, Meyer Y (1992) cDNA nucleotide sequence and expression of tobacco cytoplasmic ribosomal protein L2 gene. Nucl Acids Res 7:1517–1522
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914
Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10:11
Pfaffl MW (2004) Quantification strategies in real-time PCR. In: Bustin SA (ed) A-Z of quantitative PCR. International University Line (IUL), La Jolla, pp 87–112
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determinant of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—excel-based tool using pair-wise correlations. Biotech Let 26:509–515
Radchuk V, Sreenivasulu N, Blume Y, Weschke W (2008) Cloning and expression of the tubulin genes in barley. Cell Biol Int 32:557–559
Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A (2004) Guideline to reference gene selection for quantitative real-time PCR. Bioch Biophys Res Com 313:856–862
Rapacz M, Kościelniak J, Jurczyk B, Adamska A, Wójcik M (2010) Different patterns of physiological and molecular response to drought in seedlings of malt and feed- type barleys (Hordeum vulgare). J Agron Crop Sci 196:9–19
Rasoulnia A, Bihamta MR, Peyghambari SA, Alizadeh H, Rahnama A (2010) Proteomic response of barley leaves to salinity. Mol Biol Rep. doi:10.1007/s1103301006518
Schmidt GW, Delaney SK (2010) Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol Genet Genomics 283:233–241
Singh R, Green MR (1993) Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science 259:365–386
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 RESEARCH0034
Vandesompele J, Kubista M, Pfaffl MW (2009) Reference gene validation software for improved normalization. In: Logan J, Edwards K, Saunders N (eds) Real-time PCR: current technology and applications. Caister Academic Press, London, pp 47–64
Vinocur B, Altman A (2005) Recent advantages in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotech 16:123–132
Wan H, Zhao Z, Qian Ch, Sui Y, Malik AA, Chen J (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399:257–261
Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9:244–252
Wardlaw IF (1990) The control of carbon partitioning in plants. New Phytol 116:341–381
Xiong L, Zhu JK (2001) Abiotic stress signal transduction in plants: molecular and genetic perspectives. Physiol Plant 112:152–166
Acknowledgments
The authors are indebted to Agata Stajuda (Edinburgh Napier University) for critical reading of the manuscript. This work was supported by the European Regional Development Fund through the Polish Innovative Economy Program 2007–2013 project WND-POIG.01.03.01-00-101/08 POLAPGEN-BD “Biotechnological tools for breeding cereals with increased resistance to drought”.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Sowinski.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Rapacz, M., Stępień, A. & Skorupa, K. Internal standards for quantitative RT-PCR studies of gene expression under drought treatment in barley (Hordeum vulgare L.): the effects of developmental stage and leaf age. Acta Physiol Plant 34, 1723–1733 (2012). https://doi.org/10.1007/s11738-012-0967-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11738-012-0967-1