Abstract
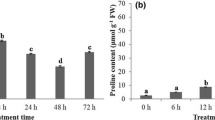
Drought is one of the major abiotic stresses limiting crop productivity in arid and semi-arid regions and influences many aspects of plant development. Groundnut (Arachis hypogaea L.) is an important oil yielding crop and considered as relatively drought tolerant. In this study, two groundnut cultivars were first tested for their drought tolerance based on physiological marker attributes such as relative water content, total chlorophyll content, cell membrane stability and free proline content and identified cultivar K-134 as a drought tolerant and cultivar JL-24 as drought susceptible. To gain a better understanding of the drought stress responses at molecular level, we carried out a genomic analysis of stress-responsive genes/transcripts in drought-tolerant cultivar K-134. As a first step toward characterization of stress-responsive genes, construction and analysis of subtracted cDNA library from drought-tolerant cultivar (K-134) is reported here. Using this strategy a total of 200 ESTs were isolated, sequenced, of which 120 high-quality ESTs were obtained and clustered. Further, our analysis revealed that 31% sequences were unique and no homology to known proteins in the database. This observation has great relevance since groundnut is a stress-adapted legume crop. Further, to validate the identified differentially expressed genes, expression profiles of selected clones were analyzed using dot blot (reverse northern), northern blot analysis. We showed that these clones are differentially expressed under different abiotic stress conditions. The implications of the analyzed genes in abiotic stress tolerance were discussed.





Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zang J, Zang Z, Miller W, Lipman DJ (1997) Gapped BLASTA and PSI-BLASTA: a new generation of protein data base search programmes. Nucleic Acids Res 25:1042–1054
Arnon DI (1949) Copper enzyme in isolated chloroplast poly phenol oxidase in Beta vulgaris. Plant Physiol 24:1–15
Arunyanark A, Jogloy S, Akkasaeng C, Vorasoot N, Kesmala T, Nageswara Rao RC, Wright GC, Patanothai A (2008) Chlorophyll Stability is an Indicator of Drought Tolerance in Peanut. J Agro Crop Sci 194(2):113–125
Bajaj S, Targolli J, Liu LF, Ho THD, Wu R (1999) Transgenic approaches to increase dehydration stress tolerance in plants. Mol Breed 5:493–503
Bajji M, Lutts S, Kinet JM (2001) Physiological changes after exposure to and recovery from polyethylene glycol-induced water deficit in callus cultures issued from durum wheat (Triticum durum Desf.) cultivars differing in drought resistance. J Plant Physiol 156:75–83
Barrs HD, Weatherly PE (1968) A re-examination of the relative turgidity for estimating water deficits in leaves. Aust J of Biol Sci 15:413–428
Bartels D, Hanke C, Schneider K, Michel D, Salamini F (1992) A desiccation-related Elip-like gene from the resurrection plant Craterostigma plantagineum is regulated by light and ABA. EMBO J 11:2771–2778
Bates LS, Waldren RP, Teare ED (1973) Rapid determination of free proline for stress studies. Plant Soil 39:205–208
Blomstedt CK, Gianello RD, Gaff DF, Hamill JD, Neale AD (1998) Differential gene expression in desiccation-tolerant and desiccation-sensitive tissue of the resurrection grass (Sporobolus stapfianus). Aust J Plant Physiol 25:937–946
Bomati EK, Noel JP (2005) Structural and kinetic basis for substrate selectivity in Populus tremuloides sinapyl alcohol dehydrogenase. Plant Cell 17:1598–1611
Boominathan P, Shukla R, Kumar A, Manna D, Negi D, Verma PK, Chattopadhyay D (2004) Long term transcript accumulation during the development of dehydration adaptation in Cicer arietinum. Plant Physiol 135:1608–1620
Chimenti CA, Pearson J, Hall AJ (2002) Osmotic adjustment and yield maintenance under drought in sunflower. Field Crops Res 75:235–246
Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81:1991–1995
Cushman JC, Bohnert HJ (2000) Genomic approaches to plant stress tolerance. Curr Opin Plant Biol 3:117–124
Datta K, Schmiit A, Marces A (1989) Characterization of two Soybean repetitive proline rich proteins and a cognate cDNA from germinated axes. Plant Cell 1:9445–9952
Delauney AJ, Verma DPS (1993) Proline biosynthesis and osmoregulation in plants. Plant J 4:215–223
Duncan M (1955) Multiple range and multiple tests. Biometrics 42:1–47
Espartero J, Pintor-Toro JA, Pardo JM (1994) Differential accumulation of S-adenosyl methionine synthetase transcripts in response to salt stress. Plant Mo1 Biol 25:217–227
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Giridarakumar S, Matta Reddy A, Sudhakar C (2003) NaCl effects on proline metabolism in two high yielding genotypes of mulberry (Morus alba L.) with contrasting salt tolerance. Plant Sci 165:1245–1251
Gopalakrishna R, Ganesh K, Krishna Prasad BT, Mathew MK, Udaya Kumar M (2001) A stress-responsive gene from groundnut, Gdi-15, is homologous to flavonol 3-o-glucosyltransferase involved in anthocyanin biosynthesis. Biochem Biophys Res commun 284:574–579
Grimm B, Kruse E, Kloppstech K (1989) Transiently expressed early light-inducible proteins share transmembrane domains with light harvesting chlorophyll-binding proteins. Plant Mol Biol 13:583–593
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Phys Plant Mol Biol 51:463–499
Huner NPA, Oquist G, Sarhan F (1998) Energy balance and acclimation to light and cold. Trends Plant Sci 3:224–230
Hutin C, Nussaume L, Moise N, Moya I, Kloppstech K, Havaus M (2003) Early light-induced proteins protect Arbidopsis from Photooxidative stress. Proc Natl Acad Sci USA 100:4921–4926
Jing Z, De-Xiang D, Quan-Hong Y, Jian Z, Fei X, Jian-Min C, Ai-Sheng X (2010) Plant Growth Regul 62:51–58
Kariola T, Brader G, Helenius E, Li J, Heino P, Palva ET (2006) Early responsive to dehydration 15, a negative regulator of ABA-responses in Arabidopsis. Plant Physiol 142:1559–1573
Kavikishor PB, Hong Z, Miao G, Hu CA, Verma DPS (1995) Overexpression of Delta1-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol 108:1387–1394
Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130:2129–2141
Leopold AC, Musgrave ME, Williams KM (1981) Solute leakage resulting from leaf desiccation. Plant Physiol 68:1222–1225
Maria S, Agnes L, Zsuzsa H, Denes D, Janos G (2005) Experimental system for studying long-term drought stress adaptation of wheat cultivar. Acta Biologia Szeged 49:51–52
Mishra RN, Ramesha A, Kaul T, Nair S, Sopory SK, Reddy MK (2005) A modified cDNA subtraction to identify full-length differentially expressed genes from any given system: an alternate to DNA chip technology. Anal Biochem 345:149–159
Mohammad A, Vimlendu B, Sinha R, Singh K, Sivalingam A, Veena P, Zakwan A (2010) Isolation of cold stress-responsive genes from Lepidium latifolium by suppressive subtraction hybridization. Acta Physiol Plant 32:205–210
Montane MH, Kloppstech K (2000) The family of light harvesting regulated proteins (LHCs, ELIPs, HLIPs): was the harvesting of light their primary function. Genetics 258:1–8
Montane MH, Dreyer S, Triantaphylides C, Kloppstech K (1997) Early light inducible proteins during long term acclimation of barley to photo oxidative stress caused by light and cold-high level of accumulation by post transcriptional regulation. Planta 202:293–302
Obulreddy PC, Sairanganayakulu G, Thippeswamy M, Sudhakar Reddy P, Reddy MK, Sudhakar Chinta (2008) Identification of stress-induced genes from the drought tolerant semi-arid legume crop horsegram (Macrotyloma uniflorum (Lam.) Verdc.) through analysis of subtracted expressed sequence tags. Plant Sci 175:372–384
Ramanjulu S, Sudhakar C (1997) Drought tolerance is partly related to amino acid accumulation and ammonia assimilation: a comparative study in two mulberry genotypes differing in drought sensitivity. J Plant Physiol 150:345–350
Ramanjulu S, Sudhakar C (2000) Proline metabolism during dehydration in two mulberry genotypes with contrasting drought tolerance. J Plant Physiol 157:81–85
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual (2nd edition). Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Sci 270:467–470
Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji T, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31:279–292
Sunkar R, Bartels D, Kirch HH (2003) Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J 35:452–464
Tyagi W, Rajagopal D, Singla-Pareek SL, Reddy MK, Sopory SK (2005) Cloning and Regulation of a Stress-regulated Pennisetum glaucum Vacuolar ATPase c Gene and Characterization of its Promoter that is Expressed in Shoot Hairs and Floral Organs. Plant Cell Physiol 8:1411–1422
Valliyodan B, Nguyen HT (2006) Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol 2:189–195
Vincent D, Lapierre L, Pollet B, Cornic G, Negroni L, Zivy M (2005) Water deficits affect caffeate O-methyltransferase, lignification and related enzymes in maize leaves: a proteomic investigation. Plant Physiol 137:949–960
Wang W, Vinocur B, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic response. Trends Plant Sci 9:244–252
Weng JK, Li JX, Stout Chapple C (2008) Independent origins of syringyl lignin in vascular plants. Proc Natl Acad Sci USA 10522:7887–7892
Wierstra I, Kloppstech K (2000) Differential effects of methyl jasmonates on the expression of the early light inducible proteins and other light regulated genes in barley. Plant Physiol 124:833–844
Wong CE, Li Y, Labbe A, Guevara D, Nuin P (2006) Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol 140:1437–1450
Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14:165–183
Yoshiba Y Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Harada Y, Shinozaki K (1995) Correlation between the induction of a gene for delta 1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J 7:751–760
Zeng Q, Chen X, Wood AJ (2002) Two early light-inducible protein (Elip) cDNAs from the resurrection plant Tortula ruralis are differentially expressed in response to desiccation, rehydration, salinity, and high light. J Exp Bot 53:1197–1205
Zhang JX, Klueva NY, Wang Z, Wu R, Ho TH, Nguyen HT, Ho THD (2000) Genetic engineering for abiotic stress resistance in crop plants. In Vitro Cell Dev Biol 36:108–114
Zhuang J, Peng RH, Cheng ZM, Zhang J, Cai B, Zhang (2009) Genome-wide analysis of the putative AP2/ERF family genes in Vitis vinifera. Sci Hortic 123:73–81
Acknowledgments
The studies for this publication were conducted as part of the programme ‘Biotechnology for Dry land Agriculture in Andhra Pradesh’’ with financial support of the Research and Communication Division, Ministry of Foreign Affairs, The Government of Netherlands, through APNL-BTU, Hyderabad. We would like to thank Prof. Udayakumar (University of Agricultural Sciences, Bangalore) and Dr. Francoise Cellier (France) for providing clones.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Wang.
Rights and permissions
About this article
Cite this article
Ranganayakulu, G.S., Chandraobulreddy, P., Thippeswamy, M. et al. Identification of drought stress-responsive genes from drought-tolerant groundnut cultivar (Arachis hypogaea L. cv K-134) through analysis of subtracted expressed sequence tags. Acta Physiol Plant 34, 361–377 (2012). https://doi.org/10.1007/s11738-011-0835-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11738-011-0835-4