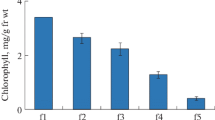
Abstract
A proline iminopeptidase (EC. 3.4.11.5) was isolated from shoots of 3 day old seedlings. The purification procedure consisted of 5 steps: acid precipitation, gel filtration on Sephadex G-200, ion-exchange chromatography on Sepharose CL 6B, twice repeated hydrophoic chromatography on Phenyl-Sepharose HP. The enzyme was purified 404.8-fold, with the specific activity of 8.5 units mg−1 of protein with recovery yield of 3%. The purified enzyme had a molecular mass of 225 kDa estimated by gel filtration and 55.4 kDa by SDS PAGE. This indicates that native enzyme is composed of four subunits. The enzyme was specific for proline β-naphtylamide among various amino acid β-naphtylamides.
An optimal activity was observed at 37 °C at pH 7.75. The enzyme was thermostable up to 37 °C for 30 min. The enzyme was strongly inhibited by pHMB, E-64, heavy metal ions and partially by PMSF, DFP. The results suggest that cysteine and serine residues may participate in the enzyme activity.
Similar content being viewed by others
Abbreviations
- 2-ME:
-
2-mercaptoethanol
- PAGE:
-
polyacrylamide gel electrophoresis
- SDS-PAGE:
-
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- EDTA:
-
ethylenediaminetetraacetic acid
- E-64:
-
transepoxysuccinyl-L-leucyloamido-(4-guanidino)butane
- pHMB:
-
p-hydroxymercuribenzoic acid
- βNA:
-
β-naphthylamide
- PMSF:
-
phenyl methylsulfonyl fluoride
- DFP:
-
diisopropyl fluorophosphate
References
Atlan D., Gilbert C., Blanc B., Portalier R. 1994. Cloning, sequencing and characterization of the pepIP gene encoding a proline iminopeptidase from Lactobacillus delbrueckii subsp. bulgaricus CNRZ 397. Microbiology 140: 527–535.
Bradford M.M. 1976. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 72: 248–254.
Blum H., Baier H., Gross H.J. 1978. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8: 93–99.
Caldwell J., Sparrow L. 1990. Purification and characterization of an unusual aminopeptidases from pea seeds. Aust. J. Plant Physiol. 7: 131–140.
Casano L.M., Desimone M., Trippi V.S. 1989. Proteolytic activities and alkaline pH oats leaves, isolation of an aminopeptidase. Plant Physiol. 91: 1414–1418.
Couton J.M., Sarat G., Wagner F.W. 1991. Purification and characterization of a soybean cotyledon aminopeptidase. Plant Sci. 75: 9–17.
Delauney A.J., Verma D.P.S. 1993. Proline biosynthesis and osmoregulation in plants. Plant J. 4: 215–223.
Dubey R.S., Rani M. 1990. Influence of NaCl salinity on the behavior of protease, aminopeptidase and carboxypeptidase in rice seedlings in relation to salt tolerance. Aust. J. Plant Physiol. 30: 133–145.
Eckey-Kaltenbach H., Ernst D., Heller W., Sanderman H. 1994. Biochemical plant responses to ozone IV. Cross-induction of defensive pathways in parsley (Petroselinum crispum L.). plants. Plant Physiol. 1041: 67–74.
Gu Y.Q., Walling L.L. 2002. Identification of residues critical for activity of the wound-induced leucine aminopeptidase (LAP-A) of tomato. Eur. J. Biochem. 269: 1630–1640.
Inoue T., Ito K., Tozaka T., Hatakeyama S., Tanaka N., Nakamura K.T., Yoshimoto T. 2003. Novel inhibitor for prolyl aminopeptidase from Serratia marcescens and studies on the mechanism of substrate recognition of the enzyme using the inhibitor. Arch. Biochem. Biophys. 416:147–154.
Isola M.C., Franzoni L. 1996. Aminopeptidase activities in peanut cotyledons. R. Bras. Fisiol. Veg. 8: 167–173.
Laemmli U.K. 1970. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227: 680–685.
Lineweawer M., Burk D.J. 1934. The determination of enzyme dissociation constans. J. Am. Chem. Soc. 56: 658–666.
Kitazono A., Kabashima T., Huang H.S., Ito K., Yoshimoto T. 1996. Prolyl aminopeptidase gene from Flavobacterium meningosepticum: Cloning, purification of the expressed enzyme, and analysis of its sequence. Arch. Biochem. Biophys. 336: 35–41.
Kolehmainen L., Mikola J. 1971. Partial purification and enzymatic properties of aminopeptidase from barley. Arch. Bioch. Biophys. 145: 633–642.
Mathushima M., Takahasi T., Ichinose M., Miki K., Kurokawa K., Tabakashi K. 1991. Prolyl aminopeptidase from pig intestinal mucosa and human liver: purification, characterization and possible identity with leucyl aminopeptidase. Biomed. Res. 12:323–33.
Medrano F.J., Alonso J., García J.L., Romero A., Bode W., Gomis-Rüth F.X. 1998. Structure of proline iminopeptidase from Xanthomonas campestris pv. citri: a prototype for the prolyl oligopeptidase family. EMBO J. 17: 1–9.
Ninomiya K., Kawatani K., Tanaka S. 1982. Purification and properties of a proline aminopeptidases from apricot seeds. J. Biochem. 92: 413–421.
Oviedo Ovando M.E., Isola M.C., Maldonado A.M., Franzoni L. 2004. Purification of properties of iminopeptidase from peanut seeds. Plant Science 116: 1143–1148.
Schaller A. 2004. A cut above the rest: the regulatory function of plant proteases. Planta 220: 183–197.
Simpson D.J. 2001. Proteolytic degradation of cereal prolamins — the problem with proline. Plant Science 161: 825–838.
Taylor A. 1993. Aminopeptidases: structure and function FASEB J. 7: 290–298.
van der Hoorn R.A.I., Jones J.D. 2004. The plant proteolytic machinery and its role in defense. Curr. Opin. Plant Biol. 55: 555–590.
van der Valk, H.C.P.M. van Bentum M.I.A., van Loon L.C. 1989. Proteolytic enzymes in developing leaves of oats (Avena sativa L.). II. Aminoacyl-2-naphtylamidases. J. Plant Physiol. 135: 489–494.
Varshavsky A., Byrd C. 1997. Recent studies of a N-end rule pathway FASEB J. 11: 1067–75.
Walling L. 2006. Recycling or regulation? The role of amino-terminal modifying enzymes. Curr. Opin. Plant Biol. 9: 227–233.
Waters S.P., Dalling M.J. 1983. Isolation and some properties of an iminopeptidases from the primary leaf of wheat (Triticum aestivum L.). Plant Physiol. 73: 1048–1054.
Yamaoka Y., Takeuchi M., Marohashi Y. 1994. Purification and partial characterization of an aminopeptidases from mung bean cotyledons. Physiol. Plant. 90: 729–733.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Szawłowska, U., Prus, W. & Bielawski, W. The molecular and biochemical characteristics of proline iminopeptidase from rye seedlings (Secale cereale L.). Acta Physiol Plant 28, 517–524 (2006). https://doi.org/10.1007/s11738-006-0047-5
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s11738-006-0047-5