Abstract
Anthracnose is one of the most relevant diseases of mango crops in producing regions, affecting 60% of production. Currently, its detection is carried out in late stages by human visual inspection. Hyperspectral imaging systems allow the development of non-destructive solutions to inspect and detect internal damage. This work aimed to develop a system for detecting anthracnose in mango fruits using Vis–NIR hyperspectral imaging and discriminant analysis. The usefulness of three-dimensionality reduction methods to minimise redundancy in the spectral data and to obtain a compact number of wavelengths that effectively allow the detection of anthracnose symptoms in mango fruits is also explored. As a result, a classification model based on discriminant analysis and Pearson correlation coefficient was obtained, showing the potential of hyperspectral data to robustly allow the detection of anthracnose symptoms with full or reduced spectra. The findings reported in this study can serve as the basis for developing an anthracnose detection system in mango fruits with multispectral cameras.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With the development of machine vision systems and the rise of specialised sensors such as multispectral and hyperspectral cameras, industry, and especially agribusiness, has undergone a technological change in the way food is produced thanks to the use of vision systems capable of performing automatic quality controls on harvested agri-food products. In recent years, the literature has reported that hyperspectral technology has shown sufficient potential for developing inspection, characterisation and disease control tools for agricultural crops [1,2,3,4,5,6]. This technology has been widely used for the non-destructive estimation of internal nutrients and the quality control of fruits, such as persimmon, bananas, strawberries, and aonla [1, 5, 7,8,9,10].
Likewise, in mango, this technology has been used for the estimation of internal fruit parameters, such as titratable acidity (TA), firmness (F) and total soluble solids (TSS), and the definition of a ripening index (RPI) [1, 2, 4]. For example, Vélez-Rivera et al. [11] presented a study on the non-invasive estimation of F, TSS and TA in Manila mango. The authors built a ripening classification model involving fruit colour parameters and a combined principal component analysis (PCA) and multivariate discriminant analysis (MDA) model. As a result, the authors obtained an accuracy of 97.44% by combining PCA and MDA. Similarly, in [12] an internal quality index (IQI) and a RPI for the Osteen mango were proposed based on near-infrared (NIR) and visible (Vis) spectroscopy. The physical and chemical properties analysed in this study were F, the colour of the pulp and peel, TA and TSS. Results of these works were confirmed in [13], where a study of the F, TA and TSS properties with hyperspectral images was carried out, achieving a strong relation of F with some spectral bands, as well as a strong correlation between F and TA. Likewise, the conclusions stated the potential of these images in different non-destructive applications for detecting mango properties (F, TA and ripeness) or damage.
Particularly, in the detection of relevant diseases in mango, anthracnose appears as the disease with the highest incidence and presence in this crop due to the difficulty for adequate detection both in the field and in the harvested fruits [14,15,16]. Studies related to anthracnose disease detection in mangoes are presented in [17, 18]. Khan et al. [17] proposed a support vector machine (SVM) model to classify anthracnose lesions through the analysis of colour images of mango leaves and their correlation with chlorophyll and carotenoids, while [19] presented a multilayer convolutional neural network (MCNN) trained on a bank of 2200 colour images taken from anthracnose-infected mango samples. Similarly, in [18], a pre-trained MobileNetV2+SVM model was used to identify and classify anthracnose disease, bacterial black spot, and sooty mold in mango using 380 colour images representative of healthy and diseased classes. To improve generalisation and the learning stage, different data augmentation techniques were applied. Likewise, Ramirez et al. [20] proposed a linear discriminant analysis (LDA) model to detect anthracnose in Sugar mango fruits using colour and ultraviolet (UV) images. The authors stated the feasibility of detecting the disease at early stages using UV-A light and the potential of LDA to discriminate mangoes infected with anthracnose. Despite the results and advances reported in these works, all models were based on the use of colour technology mimicking the human eye, which implies that the detection of the disease can only be done at a visible or late stage, when the disease can be spread to other fruits.
The aim of this study was focused on the use of hyperspectral imaging to detect anthracnose symptoms in mango Kent fruits through the implementation and assessment of dimensionality reduction techniques and DA-based classification models. The study starts with selecting and inoculating mango fruit in controlled conditions with Colletotrichum gloeosporioides sp., followed by hyperspectral image acquisition with a laboratory hyperspectral imaging system. Following the acquisition, image correction procedures, data filtering, selection of regions of interest (ROIs), and data labeling were carried out to define the control and inoculated (diseased) classes. Subsequently, through dimensionality reduction techniques (Pearson’s correlation coefficient, Tukey’s test and PCA) and DA-based models (LDA, quadratic discriminant analysis—QDA and partial least squares-discriminant analysis—PLSDA), a comparative analysis of the obtained results was performed in order to assess (1) the feasibility of reducing the number of relevant (independent) wavelengths and (2) the potential of DA-based models for the temporal detection of symptoms associated with anthracnose in mango. As a main outcome, it demonstrated the viability of developing a robust spectral vision system for detecting anthracnose in mango using DA-based classification models and reducing the number of wavelengths needed to reliably discriminate between the different stages of disease development.
This work is organised as follows. Section “Materials and methods” introduces the materials and methods used in this study, including the fruit selection, the inoculation procedure, the hyperspectral acquisition system, the dimensionality reduction techniques and the DA-based classification models. Section “Results and discussion” presents the results obtained with the DA-based classification models using the full spectrum and the reduced group of wavelengths extracted by the dimensionality reduction methods. Lastly, section “Conclusions” presents the main conclusions, outlining the novel contribution of this work and the possibilities opened by the results to deepen and extend the design of more robust spectral models.
Materials and methods
Fruit selected for the study
Sixty Kent mangoes were selected by an expert from a producing farm in Malaga (Spain) considering similar physiological maturity (colour and size) without visible lesions, see Fig. 1. Fruits were disinfected by immersion in a 0.5% hypochlorite solution for 30 s and then dried with paper towels, as reported in [21].
The fruits were separated into two groups: 10 control units and 50 units for inoculation and development of anthracnose symptoms. On all fruits, six wounds were made with a surgical punch on one side of the fruit, close to the equatorial zone, this area was selected taking into account the curvature (almost null) of the fruit and the sufficient space to capture in a single hyperspectral image different symptoms caused by anthracnose. Lesions of the control group fruits (knc) were covered with distilled water, whilst the lesions of the inoculated group (kni) were covered with a solution of Colletotrichum gloeosporioides sp. inoculum at a concentration of 1 × 106 conidia/ml. All fruits were then stored in a thermally controlled chamber at 23 °C and relative humidity > 85% for 12 days from day 1 of inoculation. The control and inoculated groups were spatially separated to avoid cross-contamination between the two groups and to prevent pathogen spores from infecting samples in the control group.
Hyperspectral acquisition vision system
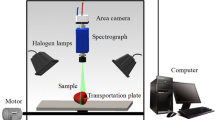
Forty-eight hours after fruit inoculation, hyperspectral image acquisition was started for 8 days on all fruits with a daily acquisition frequency. The hyperspectral acquisition system consists of a hemispherical dome with 12 halogen lamps, a monochromatic camera (Photometrics CoolSnap ES) with sensitivity in the range 320–1030 nm and two liquid crystal tunable filters (LCTF) in the visible (450–720 nm) (Varispec VIS07) and near-infrared (730 and 980 nm) (Varispec NIR07) range (Fig. 2a). Throughout the acquisition of hyperspectral images, the fruits were placed on a motorised elevator that positioned them at 14 cm from the filter to ensure uniformity and repeatability of the recorded fruit face area. Additionally, white (99% Spectralon reference panel) and black (filters covered, dark chamber and lamps off) reference images were taken at the beginning and end of each session, respectively. A total of 480 hyperspectral images were collected: 80 hyperspectral images for the control group and 400 for the inoculated group.
To acquire the hyperspectral images, an estimated time of 3 h per day was employed. Most of this time corresponded to the acquisition time of the fruit images and calibration references (2 min per image, resulting in an estimated 125–130 min), turning on and stabilisation of the light source (10–15 min approximately) and the disinfection of the hemispherical dome to avoid cross-contamination between kni and knc groups.
Reflectance correction and ROI selection
The relative reflectance of the fruits (\({I}_{ref}\)) was obtained by correcting the acquired image (\({I}_{raw}\)) with white (\({W}_{ref}\)) and black (\({B}_{ref}\)) references using Eq. (1). This procedure corrects the reflectance range associated with each fruit and avoids misinterpretations of the spectral signatures associated with the scene (fruits) captured. After this processing, the background of the scene was removed by binary thresholding at wavelength 700 nm.
The selection of the regions of interest (ROI) was performed on the six lesions caused on each fruit by the surgical punch (red inner contours in Fig. 2b); therefore, for each fruit and on each day, 6 ROI were selected. From the last day of acquisition (day 9), circular regions (cyan-coloured regions in Fig. 2b) with a radius between 3 and 5 mm for the inoculated group (kni) and 5 mm for the control group (knc) were extracted around each surgical lesion, considering that each ROI covered the size of the developed anthracnose lesion in the fruits of the kni group and a visibly sound portion (without anthracnose-associated symptoms) in the fruits of the knc group. Then, the 6 ROIs from day 9 were projected to the rest of the images (day 2–8) to ensure that the size of the ROI represented the same area on the fruit during each day of acquisition. That is, for the kni group, each ROI represents the temporal development of anthracnose symptoms in a specific region of the fruit, while for the knc group, each ROI depicts changes related to the natural ripening process of the fruit without the development of any pathological lesions. With this strategy, 2880 ROI were extracted: 480 ROI for the knc group and 2400 for the kni group. However, given the high volume of spectral signatures per ROI (820 spectral signatures), each ROI was sub-sampled by calculating the average signature of a 3 × 3 pixel window on each region. The size of this window was selected considering the size of a detectable anthracnose lesion (> 1 mm). With this sub-sampling, 252,801 spectral signatures distributed in nine classes were obtained. These classes were randomly divided into training (70% of the spectral signatures) and test (30% of the spectral signatures) groups, see Table 1.
Finally, the classes were established considering the group and the day after inoculation (dai). Since the study aimed to characterise the development of anthracnose from an early (predictable) to a visible (late) stage, the spectral signatures of kni were divided by dai. On the other hand, since the fruits of knc did not show any visible lesions associated with diseases or external agents, the control class was formed with the extracted signatures from all ROI in this group. In Fig. 3 is shown the spectral mean signature of the classes, and it can be noted the spectral difference of the knc group in the range 730–850 nm. This difference is most noticeable in the inoculated groups on days 7–9 (kni_d7, kni_d8 and kni_d9) when the anthracnose symptoms start to become visible and detectable by the human eye.
Discriminant analysis-based models for anthracnose detection
Discriminant analysis (DA) is a supervised technique used in spectral analysis to separate groups considering the inter and intra-group variance of the classes. Although there are other classification models not based on DA (classification and regression tree—CART, random forest—RF, Naive Bayes), a preliminary analysis of these models determined that the DA-based models were better fitted to the spectral data obtained in this study. As a reference for the comparative testing among these models, the results associated with the performance of the CART, RF and Naive Bayes models can be found in Appendix.
As DA models, LDA, QDA and PLSDA techniques were implemented and compared in this study. LDA produces linear decision boundaries that maximises the inter-group variance and minimises the intra-group variance. In contrast, QDA produces quadratic (curved) boundaries that allow to separate groups when they have non-linear relationships. Likewise, PLSDA is a robust algorithm that has proven to be very successful in modelling high-dimensional datasets for diverse purposes, for instance, product validation in food analysis and disease classification in medical diagnostics [22].
Since the data shown in Table 1 had no balanced class distribution, the prior class probabilities for the models were obtained by dividing the number of samples in each class by the total number of samples. For LDA, eight linear discriminants, LD1 to LD8, were defined and explained the 36.29%, 22.34%, 17.87%, 8.81%, 5.93%, 4.86%, 2.57%, and 1.32% of the total variance, respectively. For QDA, as decision boundaries are not linear, the model explained the total variance of the data.
Similarly, since the DA-based models estimate the parameters associated with the discriminant probability functions, the parameter computation time essentially depends on the number of data used in the training (calibration) of these models. Therefore, the estimated calibration time of the three models (LDA, QDA and PLSDA) using the training group (shown in Table 1) was 30 min running Rstudio 2022.02.3 Build 492 on a computer with a 3.2 GHz AMD Ryzen 7 processor x16, 64 GB RAM, 1 TB solid state disk and a NVIDIA GPU with 4 GB RAM. The speed at which DA-based models are calibrated provides an advantage in the implementation of spectral models since they do not use iterative processes (such as RF, artificial neural networks, SVM, among others) and do not have hyperparameters that directly affect the design and performance of the classification model.
Dimensionality reduction of variables
Dimensionality reduction techniques were used in this study to (1) reduce the redundant spectral information and (2) find a group of significant variables that allow an efficient classification of the models. The benefits and advantages of using this kind of procedure are exposed in previous works [3, 23,24,25]. While in Gómez-Sanchıs et al. [23], dimensionality reduction was used to reduce redundant information and improve the performance of the system, in Yeh et al. [3] this process was applied to select the most significant variables to classify three stages of anthracnose development in strawberry.
In this study, the variable selection was made by Pearson’s correlation coefficient (Correlation), Tukey test and PCA to get a group of important variables as inputs of the DA model training. The first method (Correlation) finds pair-wise correlations among the variables higher than a threshold (0.9 for this study) to discard redundant variables, while Tukey test finds, through a multivariate analysis, variables with means significantly different to allow classification among the classes.
Given the versatility and potential use of the PCA technique in hyperspectral analysis for dimensionality reduction, a reference study addressing different PCA-based reduction techniques was presented in [24]. However, the approach used here was the one presented in [25], as in this work PCA was applied to spectral signatures measured from a batch of mango fruits. Therefore, PCA was applied by organising the n − 1 principal components (PCs) according to the percentage of explained variance, where n is the number of wavelengths (54). The number of PCs selected for this study was 6, corresponding to 99% of the explained variance of the data. Figure 4a shows the first 10 PCs where it can be denoted that from PC 7 onwards the variance explained by these PCs is minimal (the sum is less than 1%). Then, the loadings associated with each wavelength were analysed for the six previously selected PCs (Fig. 4b). The idea of this analysis was to determine whether the loadings at each PC correspond to peaks or valleys to select them as relevant variables for the dataset.
a Percentage of variance explained for the first 10 PCs, where the first 6 PCs explain 99% of the variance of the data under analysis; and b Weight function of each of the six selected PCs, where the absolute peaks of each function establish the contribution of each wavelength to the reconstruction of each PC
The Correlation method, Tukey test and PCA obtained 27, 20 and 27 wavelengths, respectively, from the original spectral signature with 54 wavelengths (Table 2). This means that Correlation and PCA reduced the number of variables by 50%, while the Tukey test the reduction was 62.96%. As these methods do not perform a ranking or analysis of the variables that contribute most to the characterisation of the data, it was considered that the selected variables contributed equally to the description and discrimination of the defined classes.
Results and discussion
This section details the results obtained for the models and methods presented in sections “Discriminant analysis-based models for anthracnose detection” and “Dimensionality reduction of variables”. The metrics used to measure the performance of the models with the test data were accuracy, kappa, specificity, sensitivity (or recall), F1, balanced accuracy and Mathews correlation coefficient (MCC). The specificity, recall, F1 and balanced accuracy metrics represent the average of these metrics for all classes, as these metrics are calculated for binary classification and averaged for multi-class classification. Finally, the balanced accuracy and MCC metrics were included in this study to manage class imbalance and validate the results obtained with the recall, specificity and F1 metrics: a value close to 1 in these two metrics indicates that the model fits properly to the data used by the model regardless of the existing imbalance among the classes.
Dimensionality reduction results
From the group of wavelengths shown in Table 2, it can be noted that there are regions that offer a greater contribution to the discrimination between classes: 460–520 nm, 680–730 nm, 880–970 nm. In Fig. 5 it can be denoted the spectral distribution of the selected wavelengths per technique: Fig. 5a denotes the wavelengths obtained by the correlation method, Fig. 5b the wavelengths got using the Tukey’s test, while Fig. 5c shows the wavelengths selected using PCA. Furthermore, there were five selected wavelengths common to all techniques: 460, 520, 690, 760 and 950 nm, which could suggest they are relevant for the characterisation of the anthracnose symptoms. The regions 460–520 nm and 680–730 nm are associated with pigments such as chlorophyll a and b and β-carotene [17, 23]. This may be because the development of the infection in the epidermis of the fruit causes the degradation of these pigments [17], inducing the premature death of the affected tissue and, therefore, inhibiting the natural ripening process of the fruit. Furthermore, the 880–970 region may be of interest for the anthracnose detection in mango because the 880–940 nm range is closely related to biochemical and physiological changes (degradation of carbohydrates such as starches and sugars) induced by the pathogen in the fruit [26], while the 950–970 nm range is associated with an absorption peak of the water content in the fruit [23], which may indicate that the infectious process causes a rupture of the cellular tissue and, therefore, an accumulation of water under the epidermis (which causes an absorption peak).
Wavelength distribution of dimensionality reduction methods: a Pearson’s correlation coefficient; b Tukey’s test; and c PCA. The greyish regions in these figures highlight the common wavelengths for all methods, while the greenish regions denote the common wavelengths between Tukey test and PCA, the purple ones the wavelengths between Correlation and PCA, and the orange ones the wavelengths between Correlation and Tukey test (Color figure online)
However, a notable difference among the methods lies in the isolated or unique regions of each technique. In the Correlation method, it can be noted that the selection of variables was centred on the Vis range (Fig. 5a), i.e. the method focused on the external pigmentation changes of the fruit [17], which, despite being associated with anthracnose symptoms, corresponds to a late or visible stage of disease development. Despite having the same number of extracted wavelengths, the PCA method achieved a wider dispersion of the selected wavelengths as it extracted them from both spectral ranges (Vis and NIR), see Fig. 5c. Contrary to previous methods, the Tukey test technique put more emphasis on the NIR range, indicating a higher potential to discriminate non-visible symptoms of anthracnose in mango (carbohydrates and cellulose degradation) [23, 26, 27] since this region is not visible to the human eye, thus offering an excellent opportunity for the development of early detection solutions for this disease in mango.
DA-based classification models
The LDA, QDA and PLSDA models were computed with the wavelengths found by the dimensionality reduction methods and compared with the models computed with the full spectral range. Table 3 shows the results associated with each variable configuration and DA model through the performance metrics accuracy, kappa, specificity, sensitivity (or recall), F1, balanced accuracy and MCC.
These results showed that the best-performing model for each variable setting was QDA, with the QDA-full spectrum method being the best performing with an accuracy = 0.909, kappa = 0.898, specificity = 0.989, sensitivity = 0.929, F1 = 0.916, MCC = 0.899, and balanced accuracy = 0.959. This model was followed by the QDA-Correlation model with close metrics: accuracy = 0.876, kappa = 0.860, specificity = 0.985, sensitivity = 0.899, F1 = 0.885, MCC = 0.862, and balanced accuracy = 0.942, which evidenced the redundancy and poor contribution of variables (and certain regions) to discriminate among the established classes. Following the QDA model, the LDA model reached good performance with the full spectrum and correlation method configurations, while the PLSDA model had the lowest performance of all the models evaluated. Despite the class imbalance (knc versus kni classes), the kappa, balanced accuracy and MCC metrics achieved a value higher than 0.86 for these models (QDA-full spectrum and QDA-Correlation), showing that the models fitted the spectral data and not the number of observations used for training and validation: the success of DA-based models relies on the fact that they focus on finding the maximum inter-class difference while minimising the intra-class distance.
The PCA method also showed outstanding performance with the QDA model, however, none of the PCA models reached or outperformed the Correlation models despite having the same number of wavelengths, 27. This indicated that the dimensionality reduction applied by the Correlation method fitted and characterised better the spectral data under study than the PCA method. On the other hand, the Tukey test, despite having a lower number of wavelengths (20) than the other methods, achieved good performance with the QDA-Tukey model, demonstrating the potential of this technique for the development of a multispectral anthracnose detection model. While the QDA-Correlation model achieved an accuracy = 0.876 with 50% fewer variables (compared to QDA-full spectrum), the QDA-Tukey reduced the accuracy by only 0.078 with 63% fewer variables.
On the other hand, according to the confusion matrices of the QDA models shown in Fig. 6, there were two main types of error: (1) misclassification of sound (knc) as diseased (kni) samples (type error II in matrix confusion) and (2) misclassification of diseased (kni) as sound (knc) samples (type error I). The first error, shown in the first row (from column 2 and so on), was relevant in this analysis due to the considerable amount of sound samples misclassified as diseased. This situation could be explained by the amount of sound samples used in the test data, but also by the fact that spectral differences at early stages of anthracnose lesions were minimal, which confuses the DA-based models to discriminate the classes correctly. However, as the control class (knc) was compared with diseased classes on later days (kni_d2, kni_d3, kni_d4 and so on), the incidence of this error decreased considerably. Moreover, from the field of automatic inspection and quality control of fruit, this error can be considered as a warning signal to carry out a more thorough inspection of the fruit detected as possibly diseased, which does not represent any risk for the batch under inspection as the fruit could be set aside from the rest.
Confusion matrix of the a QDA-full spectrum, b QDA-Correlation, c QDA-Tukey and d QDA-PCA models for the test group shown in Table 1
The second error, type error I, shown in the first column (from row 2 and so on), was the most critical in classification as it represented the diseased samples that the model was not able to detect, leading to the spread of the pathogen and contamination of new fruits. An explanation of this error lay in the ROI strategy applied in this study. This strategy considered the size of the anthracnose lesion on day 9 to extract the samples, so the same size lesion on the previous days may have sound areas labelled as diseased. Hence, as a strategy to overcome this problem, a robust data filtering of ROI to correctly assign the class and the introduction (and combination) of different preprocessing filters such as standard normal variate (SNV), multiplicative scattering correction (MSC) or continuum removal (CR) could help to reduce the incidence of this error.
Conclusions
This study demonstrated the feasibility of anthracnose detection using a multi-class classification model based on DA. The results show that the QDA model presents the best performance for anthracnose detection in mangoes regardless of the number of variables used: accuracy = 0.909 with 54 variables (QDA-full spectrum model), accuracy = 0.876 with 27 variables (QDA-Correlation model) and accuracy = 0.798 with 20 variables (QDA-Tukey model). Furthermore, among the results associated with this model, it can be stated that hyperspectral or multispectral technology is feasible as an automated vision solution depending on the vision system required. Likewise, the QDA model showed remarkable performance in calibration time and prediction. Hence, its use (as well as DA-based models) allows the development of solutions capable of supporting real-time fruit inspection and control tasks.
On the other hand, the relevance of using dimensionality reduction methods was evidenced since they reduced by 50% or more the number of wavelengths needed to establish a classification model without significantly affecting its performance. Moreover, thanks to the use of dimensionality reduction methods, it was found that there are regions of interest and specific wavelengths that could allow the spectral characterisation of anthracnose lesions in mango fruits. These wavelengths cover from the visible range (where anthracnose symptoms are associated with pigment degradation of the fruit) to the NIR range, where anthracnose symptoms may be related to carbohydrate degradation and water release on the peel of the fruit.
Finally, as this study involved only one mango variety, future work is planned to extend this study to other mango varieties to demonstrate the feasibility of anthracnose detection regardless of the variety. Besides, a comparative study of other classification techniques, such as neural networks, decision trees or SVM is proposed to determine the existence of an anthracnose classification model with better performance than the QDA model. Likewise, feature selection techniques (Chi-squared, RF, genetic algorithm) are proposed to determine the existence of wavelengths closely correlated to anthracnose symptoms in mango fruits.
Data availability
Not applicable.
References
E. Hamuda, M. Glavin, E. Jones, A survey of image processing techniques for plant extraction and segmentation in the field. Comput. Electron. Agric.. Electron. Agric. 125, 184–199 (2016). https://doi.org/10.1016/j.compag.2016.04.024
P. Rungpichayapicheta, B. Mahayotheeb, M. Naglea, P. Khuwijitjarub, J. Müllera, Robust NIRS models for non-destructive prediction of postharvest fruit ripeness and quality in mango. Postharvest Biol. Technol. 111, 31–40 (2016). https://doi.org/10.1016/j.postharvbio.2015.07.006
Y. Yeh, W. Chung, J. Liao, C. Chung, Y. Kuo, T. Lin, Strawberry foliar anthracnose assessment by hyperspectral imaging. Comput. Electron. Agric.. Electron. Agric. 122, 1–9 (2016). https://doi.org/10.1016/j.compag.2016.01.012
J.P. dos Santos Neto, M.W. Dantas de Assis, I. Parkutz Casagrande, L.C. Cunha Júnior, G.H. de Almeida Teixeira, Determination of ‘Palmer’ mango maturity indices using portable near infrared (VIS–NIR) spectrometer. Postharvest Biol. Technol. 130, 75–80 (2017). https://doi.org/10.1016/j.postharvbio.2017.03.009
S. Munera, C. Besada, N. Aleixos, P. Talens, A. Salvador, D. Sun, S. Cubero, J. Blasco, Non-destructive assessment of the internal quality of intact persimmon using colour and VIS/NIR hyperspectral imaging. LWT 77, 241–248 (2017). https://doi.org/10.1016/j.lwt.2016.11.063
I. Navrozidis, T. Alexandridis, A. Dimitrakos, A. Lagopodi, D. Moshou, G. Zalidis, Identification of purple spot disease on asparagus crops across spatial and spectral scales. Comput. Electron. Agric.. Electron. Agric. 148, 322–329 (2018). https://doi.org/10.1016/j.compag.2018.03.035
S.E. Adebayo, N. Hashim, K. Abdan, M. Hanafi, K. Mollazade, Prediction of quality attributes and ripeness classification of bananas using optical properties. Sci. Hortic.Hortic. 212, 171–182 (2016). https://doi.org/10.1016/j.scienta.2016.09.045
C. Guo, F. Liu, W. Kong, Y. He, B. Lou, Hyperspectral imaging analysis for ripeness evaluation of strawberry with support vector machine. J. Food Eng. 179, 11–18 (2016). https://doi.org/10.1016/j.jfoodeng.2016.01.002
A. Wendel, J. Underwood, K. Walsh, Maturity estimation of mangoes using hyperspectral imaging from a ground based mobile platform. Comput. Electron. Agric.. Electron. Agric. 155, 298–313 (2018). https://doi.org/10.1016/j.compag.2018.10.021
C. Koley, A.K. Nirala, Activity assessment of anthracnose disease infected region of aonla. AIP Conf. Proc. 2352, 030013 (2021). https://doi.org/10.1063/5.0052513
N. Vélez-Rivera, J. Blasco, J. Chanona-Pérez, G. Calderón-Domínguez, M.J. Perea-Flores, I. Arzate-Vázquez, S. Cubero, R. Farrera-Rebollo, Computer vision system applied to classification of “Manila” mangoes during ripening process. Food Bioprocess Technol. 7(4), 1183–1194 (2014). https://doi.org/10.1007/s11947-013-1142-4
V. Cortés, C. Ortiz, N. Aleixos, J. Blasco, S. Cubero, P. Talens, A new internal quality index for mango and its prediction by external visible and near-infrared reflection spectroscopy. Postharvest Biol. Technol. 118, 148–158 (2016). https://doi.org/10.1016/j.postharvbio.2016.04.011
P. Rungpichayapichet, M. Nagle, P. Yuwanbun, P. Khuwijitjaru, B. Mahayothee, J. Müller, Prediction mapping of physicochemical properties in mango by hyperspectral imaging. Biosyst. Eng.. Eng. 159, 109–120 (2017). https://doi.org/10.1016/J.BIOSYSTEMSENG.2017.04.006
F. Jenny, N. Sultana, M. Islam, M.M. Khandaker, M.A. Bhuiyan, A review on anthracnose of mango caused by Colletotrichum gloeosporioides. Bangladesh J. Plant Pathol. 35(1), 65–74 (2019)
O.F. Espinel, C.A. Velasquez, F.A. Prieto, Detection of anthracnose in mango leaves by hyperspectral image analysis, in 2021 XXIII Symposium on Image, Signal Processing and Artificial Vision (STSIVA) (2021), pp. 1–6. https://doi.org/10.1109/STSIVA53688.2021.9592005
L. Zakaria, Diversity of Colletotrichum species associated with anthracnose disease in tropical fruit crops—a review. Agriculture 11(4), 297 (2021). https://doi.org/10.3390/agriculture11040297
M.S. Khan, S.B. Uandai, H. Srinivasan, Anthracnose disease diagnosis by image processing, support vector machine and correlation with pigments. J. Plant Pathol. 101, 749–751 (2019). https://doi.org/10.1007/s42161-019-00268-9
M. Prabu, B.J. Chelliah, Mango leaf disease identification and classification using a CNN architecture optimised by crossover-based levy flight distribution algorithm. Neural Comput. Appl.Comput. Appl. 34(9), 7311–7324 (2022). https://doi.org/10.1007/s00521-021-06726-9
U.P. Singh, S.S. Chouhan, S. Jain, S. Jain, Multilayer convolution neural network for the classification of mango leaves infected by anthracnose disease. IEEE Access 7, 43721–43729 (2019). https://doi.org/10.1109/ACCESS.2019.2907383
L.A. Ramirez, C.E. Cabrera, F.A. Prieto, A computer vision system for early detection of anthracnose in sugar mango (Mangifera indica) based on UV-A illumination. Inf. Process. Agric. (2022). https://doi.org/10.1016/j.inpa.2022.02.001
J.M. Tovar-Pedraza, J.A. Mora-Aguilera, C. Nava-Díaz, N.B. Lima, S.J. Michereff, J.S. Sandoval-Islas, M.P.S. Câmara, D. Téliz-Ortiz, S.G. Leyva-Mir, Distribution and pathogenicity of Colletotrichum species associated with mango anthracnose in Mexico. Plant Dis. 104(1), 137–142 (2020). https://doi.org/10.1094/PDIS-01-19-0178-RE
L.C. Lee, C.-Y. Liong, A.A. Jemain, Partial least squares-discriminant analysis (PLS-DA) for classification of high-dimensional (HD) data: a review of contemporary practice strategies and knowledge gaps. Analyst 143(15), 3526–3539 (2018). https://doi.org/10.1039/C8AN00599K
J. Gómez-Sanchıs, J. Blasco, E. Soria-Olivas, D. Lorente, P. Escandell-Montero, J.M. Martínez-Martínez, M. Martínez-Sober, N. Aleixos, Hyperspectral LCTF-based system for classification of decay in mandarins caused by Penicillium digitatum and Penicillium italicum using the most relevant bands and non-linear classifiers. Postharvest Biol. Technol. 82, 76–86 (2013). https://doi.org/10.1016/j.postharvbio.2013.02.011
J.P. Cruz-Tirado, J.M. Amigo, D.F. Barbin, S. Kucheryavskiy, Data reduction by randomisation subsampling for the study of large hyperspectral datasets. Anal. Chim. ActaChim. Acta (2022). https://doi.org/10.1016/j.aca.2022.339793
C. Cabrera Ardila, L. Ramirez Alberto, F.A. Prieto, Spectral analysis for the early detection of anthracnose in fruits of sugar mango (Mangifera indica). Comput. Electron. Agric.. Electron. Agric. 173, 1–17 (2020). https://doi.org/10.1016/j.compag.2020.105357
J. Kou, Y. Wei, X. He, J. Xu, F. Xu, X. Shao, Infection of postharvest peaches by Monilinia fructicola accelerates sucrose decomposition and stimulates the Embden-Meyerhof-Parnas pathway. Hortic. Res. (2018). https://doi.org/10.1038/s41438-018-0046-x
C.G. Sangeetha, R.D. Rawal, Nutritional studies of Colletotrichum gloeosporioides (Penz.) Penz. and Sacc. the incitant of mango anthracnose. World J. Agric. Sci. 4(6), 717–720 (2007)
Acknowledgements
This work was partially funded by the Ministerio de ciencia y tecnología de Colombia (MINCIENCIAS) through its call “convocatoria 785 para doctorados nacionales 2017”, Universidad Nacional de Colombia through its programme “convocatoria para el apoyo a proyectos de investigación y creación artística de la sede Bogotá de la Universidad Nacional de Colombia—2019” and the Sistema General de Regalías CTeI-Colombia (BPIN 2020000100415, “Desarrollo de un sistema de óptico computacional para estimar el contenido de carbono orgánico de suelos agrícolas a través de imágenes espectrales e inteligencia artificial en cultivos cítricos de Santander”, code UIS-8933) and through GVA-IVIA 52204 and GVA-PROMETEO CIPROM/2021/014.
Funding
Open Access funding provided by Colombia Consortium.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Velásquez, C., Prieto, F., Palou, L. et al. New model for the automatic detection of anthracnose in mango fruits based on Vis/NIR hyperspectral imaging and discriminant analysis. Food Measure 18, 560–570 (2024). https://doi.org/10.1007/s11694-023-02173-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11694-023-02173-3