Abstract
The identification of Stasimopus Simon, 1892 species as well as mygalomorph species has been a long-standing challenge. This is due to their conservative morphologies as well as the lack of quantifiable characters. Ocular patterns have historically been used to aid in identification, but have largely been vague and subjective. This study was the first to test for phylogenetic signal in this character to validate its use for species identification and description as well as to test the viability of it in morphospecies and species identification. The results show significant phylogenetic signal for ocular patterns in both sexes, validating its use. The results display the evolutionary change in ocular patterns across various species. Species and morphospecies show distinct clustering in morphospace, but there is overlap due to the continuous shape of the character. The methodology of applying geometric morphometrics to quantify ocular patterns can distinguish between morphospecies and shows great promise for distinguishing species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The taxonomic placement of Stasimopus, like other mygalomorph spiders, has historically been based on morphological characters. This can however be confounding as organisms may be cryptic morphologically, display high levels of homoplasy or the characters used to describe them may be vague, indiscriminate and subjective. The characters which are often used to describe mygalomorph species are: the shape of the carapace and fovea; sternum shape and the position of the sigilla; ocular patterns; chelicerae, rastellum and cuspules; the relative leg lengths and spination patterns; the pedipalp shape and relative length to the first legs; the spinnerets; and genitalia (Engelbrecht & Prendini, 2012; Jocque & Dippenaar-Schoeman, 2007). These characters are all qualitative in nature, such as relative length and shape, with no quantitative way to distinguish similar species from each other (Bond et al., 2012).
Ocular patterns are commonly used to identify spiders in most families as there is a vast amount of diversity (Crews, 2011; Polotow & Brescovit, 2014). The size and arrangement of these eyes are generally used. For this the standard measurements tend to be eye diameter across the lens, inter-eye distances and the arrangement of the different eye pairs (Crews, 2011). Eye patterns are particularly useful at the family level within the araneomorphs which show a significantly higher degree of morphological variation than the mygalomorphs. (Crews, 2011; Polotow & Brescovit, 2014). Stasimopidae typically have eight eyes, the Anterior Median eyes (AME) are known as the ‘principal’ eyes, with the other eyes (Anterior Lateral (ALE), Posterior Median (PME) and Posterior Lateral (PLE) known as ‘secondary’ eyes (Fig. 1). Principal and secondary eyes differ extensively in terms of development, structure and function (Morehouse et al., 2017). Ocular patterns are often included in species descriptions of Stasimopus where they are considered diagnostic (Engelbrecht & Prendini, 2011; Hendrixson & Bond, 2004; Hewitt, 1910; Pocock, 1902). It has however never been tested for phylogenetic signal, thus there is no hard evidence that the trait is driven by heritable change and evolution. Phylogenetic signal is based on the observation that closely related species tend to have similar morphological traits and without phylogenetic signal, these traits should not be used to identify nor describe species (Adams, 2014b; Hendrixson & Bond, 2009).
Map of the type localities and general localities of the specimens analysed. The localities are coloured according to the species found there. Numbered markers indicate type localities, unnumbered markers are specimens collected in the Karoo and assigned to species. The various shapes indicated defined locality groupings (A-F) for analyses. (A) stars, (B) squares, (C) triangles, (D) circles, (E) hexagons, (F) diamonds. Map created in QGIS version 3.4.8-Madeira (2019), available at: http://qgis.osgeo.org
Engelbrecht and Prendini (2012) used ocular patterns and described the distance of the anterior ocelli from the carapace margin, the curvature of the anterior and posterior ocular rows as well as the eye sizes relative to each other. This is a step forward in using ocular patterns as an identifiable character for Stasimopus, but is not specific enough for the fine scale variation which exists between the species. The morphological differences between the species of the genus are subtle, making the evolutionary adaptations that would have driven these differences difficult to detect. In having a better understanding of what the differences in the ocular patterns are across the genus, we may be able to make better predictions about the differing environmental pressures the species historically faced. In order to achieve this, a more quantitative methodology is required.
One solution to this is the implementation of geometric morphometric techniques. Morphometrics is a branch of comparative biology and statistics which finds methods of describing and quantifying variations of shape and relative size between organisms and the analyses make it possible to identify discrete patterns in continuous data (Hendrixson & Bond, 2009; Rohlf & Marcus, 1993; Zelditch et al., 2004c). Geometric morphometrics allows for the capturing of the entire structure being analysed in either two-dimensions or three-dimensions as coordinates of landmark points (Mitteroecker & Gunz, 2009; Rohlf & Marcus, 1993).
Morphometrics has been broadly applied to the Araneae. However, these studies tend to focus on the genitalia, carapace and legs of different spiders to identify species or genera (Bond & Beamer, 2006; Costa-schmidt & de Araujo, 2010; Fernández-Montraveta & Marugán-Lobón, 2017; Hendrixson & Bond, 2009; Prenter et al., 1995; Spasojevic et al., 2016). To the best of our knowledge no study addressed the issue of the use of morphometrics to analyses ocular patterns in the Araneae.
We aim to fill this gap and address the following two related questions: First: can we detect evolutionary signal in the ocular pattern thereby making them useful and robust identifying traits. Second, to test the viability of applying geometric morphometrics to the ocular pattern of Stasimopus species to determine their feasibility as quantitative characters to distinguish between morphospecies or possibly species.
Methods and Materials
Specimen Selection
Specimens were selected to attempt to represent as much of the Stasimopus species diversity possible. Material was loaned from various national and international collections: Albany Museum (AMGS), Grahamstown, South Africa; The National Museum (NMBA), Bloemfontein, South Africa; Ditsong Museum of Natural History (TMSA), Pretoria, South Africa; Iziko Museum of Cape Town (SAMC), Cape Town, South Africa; Museum für Naturkunde (ZMB), Berlin, Germany; National Collection of Arachnida (NCA), Pretoria, South Africa. As far as possible only holotypes, allotypes and syntype series were used, as many of the Stasimopus identifications after this are incorrect or unvalidated. Specimens collected from the Karoo area of South Africa were included in some analyses in order to test if they could be allocated to the correct species (Specimens information is available in Table S1). GPS coordinates were omitted from Table S1, as the Karoo region is of conservation concern and the IUCN conservation statuses for Stasimopus species are completely known due to the lack of data. In total 35 of the 47 currently recognised Stasimopus species were represented between the two sexes (32 species for the females; 20 species for the males) (World Spider Catalog, 2022). As well as nine previously undescribed species which are currently undergoing description.
Specimens were also split into ‘morphospecies’ according to the major biomes in South Africa (A-F in Fig. 1). Some species were also grouped due to known phylogenetic relationships.
Geometric Morphometric Data Capture
The top angle of the ocular pattern was photographed for 123 females and 66 males belonging to various species of Stasimopus. The females were photographed using a Leica M 165 C stereomicroscope attached to Leica camera (DMC-2900), whereas males were photographed on a Zeiss Axio Zoom V16 with the Axiocam 512 color and stacked with ZEN 2.3 SPI (Blue Edition). Two different systems were required as the males have a steeper carapace and thus required an imaging system with a greater depth of field. All specimens were photographed on a petri dish of glass beads for ease of adjustment, to ensure similar angles in each photograph. The specimens were selected at random when photographed to ensure that there was no operational bias in the photography process. Operational bias in this context would arise from photographing specimens of the same species directly after one another, this would lead to the operator subconsciously detecting similarities in the positions of the specimens (Bakkes, 2017; Fruciano, 2016).
Once photographed all the images were compiled using tpsUtil v1.79 (Rohlf, 2015) and were imported to tpsDig v.2.31 (Rohlf, 2015) for digitisation. Thirty-two landmarks were selected to capture as much of the shape variation as possible as seen in Fig. 2. Landmarks in the context of geometric morphometrics are distinct loci which are homologous in location between different specimens (Rodriguez-Mendoza, 2013; Seetah, 2014; Zelditch et al., 2004a). A list of operational definitions for each landmark are included in Table S2. The specimens were selected at random when digitising to ensure that there was no operational bias.
An illustration of the position of the landmarks used in the geometric morphometric analysis of Stasimopus ocular patterns. The 32 landmarks correspond with the landmark definitions given in Table S2. In all photographs, the specimen is positioned so that the chelicerae face the top of the image. The following shorthand notations are used; PLE: posterior lateral eye, PME: posterior median eye, ALE: anterior lateral eye and AME: anterior median eye
DNA Extraction, Sequencing and Alignment
Genomic DNA was extracted from the removed third right leg of each specimen. DNA extraction was performed using the Macherey-Nagel NucleoSpin® Tissue kit (Düren, Germany) following the manufacturer’s instructions.
Three gene regions were selected for sequencing to account for the differing mutational rate changes over time. These were ribosomal 16 S, mitochondrial cytochrome oxidase 1 (CO1) and nuclear elongation factor 1 gamma (EF-1ɣ). The rationale for selecting these three gene regions was that they allow for the maximisation of phylogenetically informative data at a very fine genetic scale (species level variation). Histone H3 (H3) was also sequenced but was found to not be phylogenetically informative at this level. The phylogenetic topologies of the three gene regions will show if the gene trees reflect the species tree by congruence (Doyle, 1992; Maddison, 1997).
Genomic DNA was amplified by polymerase chain reaction (PCR) for the target genes using previously published primer sequences indicated in Table S3. Amplification mixtures were prepared to reach a final volume of 50 µL containing: 2.5 mM MgCl2, 20 pmol of each primer, 10 mM dNTPs, 1 X PCR buffer, one unit of TaqDNApolymerase (Supertherm® DNA polymerase, Separation Scientific SA (Pty) Ltd, South Africa) or Emerald Amp®MAX HS PCR Mastermix (TAKARA BIO INC., Otsu, Shiga, Japan), for problematic samples as well as the EF-1ɣ gene region, in combination with 10–50 ng of extracted genomic DNA template. The PCR cycling parameters performed for the CO1 and 16 S gene regions can be viewed in Table S4, and the parameters for EF-1ɣ were as stated in Table S5. Purification of the successful amplifications was done using the Macherey-Nagel NucleoSpin® Gel and PCR Clean-up kit (Düren, Germany) according to manufacturer’s specifications. Samples which presented double bands were gel purified following the manufacturer’s specifications. The BigDye® Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, USA) was used for the cycle sequence reactions in both sequence directions. Both directions are used as a precautionary measure as this is a fine scale study where the misreading of one base pair could impact results significantly. The cycle sequencing products were precipitated using the standard protocol of sodium acetate and ethanol. All sequences generated were edited in CLC Bio Main Workbench Version 6.9 (http://www.clcbio.com). All the sequences generated from the barcoding CO1 region will be submitted to the Barcode of life database (BOLD). CO1 along with the other two gene regions were submitted to GenBank and the accession numbers are recorded in Table S1.
The CO1, 16 S and EF-1ɣ datasets were concatenated using FASconCAT v1.11 (Kück & Meusemann, 2010). The edited sequences for each gene region as well as the concatenated dataset, were aligned using MAFFT online (Katoh, 2005; Katoh & Toh, 2008). The ‘Auto’ strategy for alignment was used in MAFFT, this inspects the direction of the sequences and adjust the alignment in correlation to the first sequence.
Phylogenetic Analyses
A dated phylogeny was produced using BEAST v1.8.4 (Suchard et al., 2018). In order to determine the most appropriate fossil calibration points, numerous combinations were tested varying the fossil calibrations used. The final fossil calibration points used were as follows: the Hexathelidae fossil, Rosamygale grauvogely (Gresa-Voltzia formation, France, Triassic) (Dunlop et al., 2020; Opatova et al., 2020; Selden & Gall, 1992). Rosamygale grauvogely is thus, the first mygalomorph appearance in the fossil record, dating to 250–240 MYA (Opatova et al., 2020). The Nemesiidae fossil, Cretamygale chasei (Isle of Wight, Cretaceous) (Selden, 2002), two Ctenizidae/Halonoproctidae fossils, Ummidia damzeni and U. malonowskii (both from Baltic amber, Palaeogene) (Wunderlich, 2000). Lastly, a fossil from the family Cyrtaucheniidae, Bolostromus destructus (Dominican amber, Neogene) (Dunlop et al., 2020; Wunderlich, 1998). Each family was represented by available sequence data from GenBank for 16 S rRNA, cytochrome c oxidase 1 (CO1) and elongation factor 1 gamma (EF-1ɣ) (Table S6). Each calibration point was set by setting a hard minimum bound (the youngest possible age of the fossil based on the deposit in which it was found) and a soft upper bound (the oldest possible age).
The nucleotide substitution rate was determined using jModelTest v2.1.7 (Posada, 2008). This was done for each gene region and the best model was selected based on the Bayesian information criterion (BIC) (Posada, 2008). The results of the JModel test led to the selection of the following nucleotide substitution models; CO1: GTR + I + G (I = 0.379, G = 0.638), 16 S: GTR + G (G = 0.348), EFɣ-1: HKY + G (G = 0.299). This was used to set the prior in BEAUTi v1.8.4 (Suchard et al., 2018). Other priors set included, the rate of molecular evolution to a relaxed clock with a lognormal distribution (this allows mutational rates to vary over the tree) (Michonneau, 2017). The tree prior was set to ‘Speciation: Birth-Death Incomplete Sampling’. An uncorrelated relaxed clock was used, as this allows for each branch of the phylogeny to have a different evolutionary rate (Drummond et al., 2006). BEAST v1.8.4 (Suchard et al., 2018) was then run for 200,000,000 generations and sampled every 2000 generations. This was repeated twice for each analysis to ensure convergence. BEAST was run in conjunction with BEAGLE (Ayres et al., 2012).
Tracer v1.7.1 was used to confirm convergence (Rambaut et al., 2018). This was tested by checking the log file for each BEAST run and ensuring the ESS values were above 200. The individual runs were then compiled to ensure a normal distribution. Log Combiner v1.8.4(Suchard et al., 2018) was used to combine the multiple tree files into one file. The subsampling number was set to 50,000 and the burn-in for each tree to 50,000,000. The tree files viewed, annotated and edited in FigTree (Rambaut, 2016).
Statistical Analyses
General Statistics
All statistical analyses were performed in either MorphoJ v.1.06d(Klingenberg, 2011) or the main packages “geomorph”, “Morpho” and “GeometricMorphometricsMix”(Adams et al., 2021; Baken et al., 2021; Collyer & Adams, 2018, 2021; Schlager, 2013) in R (R Core Team, 2017).
A Procrustes fit was created using the landmarks in MorphoJ v.1.06d and R in order to superimpose all the photographs and thus remove the direct effect of size, orientation and position (Zelditch et al., 2004b, c). General procrustes analysis (GPA) was performed on all datasets before any other analyses. This was done by the ‘align principal axes’ method, which computes the average position in an iterative manner in order to minimise the Procrustes distance (Zelditch et al., 2004b). This analysis produces the distances in three-dimensional space as well as the Procrustes distances which measure shape dissimilarity. An additional output of this analysis is the centroid size for each specimen photograph.
Determining Evolutionary Relationships
Phylogenetic signal was tested for the eye patterns of the various species which had available sequence data. Strong phylogenetic signal is present if specimens that are phenotypically more similar are more closely related than specimens that are phenotypically more distinct (Adams, 2014b; Klingenberg & Gidaszewski, 2010). This is expected when organisms have experienced divergent evolution, but can be diluted by homoplasy due to convergence, parallel evolution and reversals (Klingenberg & Gidaszewski, 2010). Phylogenetic signal is tested with morphometric data, by mapping the shape data onto a known phylogeny. Permutations are then performed, simulating the null hypothesis of no phylogenetic structure by randomly assigning the various shapes to the terminal nodes of the phylogeny (Adams, 2014b; Klingenberg & Gidaszewski, 2010; Žikić et al., 2017). This was done for the male and female dataset of sequenced individuals. Phylogenetic signal was tested for average species shape. The phylogenetic signal testing conducted using the package ‘geomorph’ which is based on a Brownian motion evolutionary model. Brownian motion evolution assumes that the changes in one trait along the phylogeny is expected to have a value of zero and variance among species accumulates proportional to time (Adams, 2014b). The statistic to measure phylogenetic signal by this method is Kmult, which is equal to one under Brownian motion. A value above one would imply that more closely related taxa have a morphology in a particular trait which is more similar than expected under Brownian motion (Adams, 2014b). To account for phylogenetic uncertainty, Kmult was calculated of every tree of the posterior distribution of the afore mentioned BEAST run (Fruciano, 2016). This was done after discarding 25% of the trees as burn-in.
A Phylogenetic Generalised Least Squares (PGLS) was performed to test for evolutionary allometry in both sexes. Evolutionary allometry describes the covariation between shape and size within a phylogenetic context (Adams, 2014a). This is important in the Stasimopus genus as there is a wide variation in average species size, and size is often used to aid in species identification. Two methodologies were tested, both requiring a pruned phylogeny as well as averages for species shape and size (Adams, 2014a). The first method follows Adams (2014b) which implements a distance-based approach to PGLS in the package geomorph. The second follows Clavel et al. (2015) and is based on fitting various evolutionary models under a maximum likelihood criterion in the package mvMORPH (Clavel et al., 2015, 2019). The evolutionary models tested included Brownian motion, Ornstein-Uhlenbeck, early burst and Lamba (Clavel et al., 2015).
Partial Least Squares (PLS) is a multivariate statistical technique used to compare multiple response and explanatory variables (Pirouz, 2006; Rohlf & Corti, 2000). The morphometric implementation is 2B-PLS, this methodology finds pairs of axes in the two blocks of variables which account for the maximal covariation between the blocks (Rohlf & Corti, 2000). PLS is considered more robust than Principal Component Analysis (PCA), Canonical Variates Analysis (CVA) and alternating least squares, as model parameters do not change with new data calibrations (Pirouz, 2006). PLS is also advantages over regression as it does not assume that the variation in the one variable is caused by the other variable, but rather treats the two variables symmetrically to uncover the relationship between them (Rohlf & Corti, 2000). PLS is better at dealing with small datasets, missing data and data that is multicollinear (Pirouz, 2006). For the analyses, the first variable block was the shape data for all specimens that have genetic sequence data (after GPA), analysed as a pairwise matrix of the Procrustes distances. The second block was the pairwise distances of the various gene regions (CO1, 16 S and EF-1ɣ). These distances were obtained by using the sequences generated, and constructing the various matrices in MegaX (Kumar et al., 2018). This block was scaled as it does not share the same units as the shape variable block (Rohlf & Corti, 2000). There is no direct significance test of 2B-PLS available, thus significance was tested by permutations (1000 permutations were used). This tests the null hypothesis that there is no association between the two variable blocks (Rohlf & Corti, 2000). The strength of the covariation is measured by Escoufier RV, which is the multivariate equivalent of the correlation coefficient (Rohlf & Corti, 2000).
Determining if Identification is Possible
A Principal Component Analysis (PCA) was performed on the whole dataset to test the patterns of covariation of the landmark positions of the eye patterns. The PCA was constructed to provide a graphical representation of the variation in the data.
Regression analyses can be inflated by the multicollinearity of variables within a model (two or more dependent variables being correlated with one another), therefore Variance Inflation Factor (VIF) was used to test if this occurred in the models used in the analysis by testing the correlation and strength between dependent variables (Forthofer et al., 2007). A value of 1 indicates no correlation between variables, 1–5 indicates moderate correlation (not enough to influence results) and > 5 indicates a severe correlation which is likely to obscure results such as p-values(Ringle et al., 2018). This was tested in R with the packages “car” (Fox & Weisberg, 2019) and “DAAG” (Maindonald & Braun, 2020). This was tested between centroid size and locality and species separately (as these variables were never run in the same model as they are correlated).
A regression of size and shape was performed in order to quantify the effects of allometry (Klingenberg, 2016). This is important to take into account as real differences in shape between species or populations can be confounded with shape differences due to size (Nakagawa et al., 2017). A regression separates the component of variation of the dependent variable (shape) selected which is predicted by the independent variable (size) (Rodriguez-Mendoza, 2013). A procrustes regression with residual randomisation was performed in ‘geomorph’. A permutation test was included to test the null hypothesis that shape and size are independent for 10,000 runs. It is possible to separate the component of size which is correlated with shape; this was decided against as size is an important predictor of species in Stasimopus. As Stasimopus species get larger, the eye pattern is more stretched over the carapace, making the ocular pattern sparser by increasing the distance between the various eyes. This means that the size of the specimen directly impacts the shape of the ocular pattern. Regressions were also performed pooled by species and by locality.
Procrustes ANOVA was used to test if there was a significant relationship between procrustes distances (shape) and multiple dependent variables (Goodall, 1991). In this study the null hypothesis was rejected at a significance level of 0.05. Procrustes ANOVA was used to test for a significant relationship between shape and centroid size, species and locality for both sexes separately.
Canonical Variates Analysis (CVA) is an exploratory analysis which generates Mahalanobis distances between groups based on centroids (McKeown & Schmidt, 2013). This produces canonical variates (CVs) from the scaling and rotation of the centroids, thus showing the distances between groups (Manthey & Ousley, 2020; McKeown & Schmidt, 2013). A CVA was conducted for species and locality for both the male and female datasets in R.
Outline files were created to be able to visualise the shape changes between different principal components and discriminant factors. For this tpsDig and MorphoJ were used. The males and females were analysed separately due to a high degree of sexual dimorphism.
Pilot Study for Error Testing
The first pilot study was to test if juveniles could be included in order to increase the sample size. It was found that juveniles experience allometric growth. The ocular pattern tends to be more bulbous in juvenile samples when compared to the adult counterparts. Juvenile specimens of the genus are often left unidentified as the juvenile traits are not well documented. It was therefore decided to exclude juveniles from the study as it would further complicate the results and take away from the straightforward testing if the method can be used to aid in the identification species. A second pilot study was conducted to determine the amount of rotational and digitising error that was incurred and the number of replications that would be required to reduce this. The pilot study found that for the eye patterns there was less than 5% total error when two replications were used. For this reason, only one replicate was performed in the actual experiment. The documentation of this is available in Appendix A.
Results
Phylogenetic Signal and Relationships
The relative position and size of the Stasimopus eyes display significant phylogenetic signal for both the female and male dataset (Kmult=1.0763; Prand=0.007 and Kmult=1.0121; Prand=0.011 respectively). This indicates that closely related species are significantly similar in eye pattern than what would be expected under a Brownian motion evolutionary model. The Kmult distribution is shown in Figure S1. This is supported by the positions of the various species in phylomorphospace being reflective of the evolutionary relationships shown in Fig. 4 (Fig. 3). Here sister species cluster closer together in morphospace than more distantly related species, indicating that they are more morphologically similar. Figure 4 also displays the phylogenetic relationships between the various species, with the average species eye pattern. From this it is clear that more closely related species have more similar eye patterns.
Phylomorphospace plots of the first two principal components for the female and male datasets. Species colours correspond to the map in Fig. 1
Molecular phylogeny of the sequenced Stasimopus specimens. Species colours correspond to the map in Fig. 1. The average shape based on thin plate splines averaged by species is given for each sex when available
The results of the PGLS analyses yielded significant results for the female datasets following both the method applied in geomorph and mvMORPH (Lamba model) (p = 0.033; p = 0.001), indicating that evolutionary allometry is present. For the male dataset evolutionary allometry was not found under geomorph but a significant result was found through mvMORPH (Early burst model) (p = 0.094; p = 0.001).
The PLS analyses for the female sequenced dataset produced significant results for all three gene regions, indicating that there is an association between the genetic distances and Procrustes distances. The Escoufier RV is however fairly low, thus the correlation is not strong (CO1: p = 0.001, RV = 0.43; 16 S: p = 0.001, RV = 0.36; EF-1ɣ: p = 0.001, RV = 0.37). For the male dataset, only 16 S and EF-1ɣ gave significant results, both with average correlation (16 S: p = 0.008, RV = 0.47; EF-1ɣ: p = 0.02, RV = 0.55). The figures showing the separation of species in the PLS blocks are available in Figures S2 and S3.
Separation of Species or Morphospecies
Eleven PCs explain 87% of the total variation in the female dataset with PC1 and PC2 explaining 20.79% and 20.15% respectively (Figure S4). For the males nine PCs explain 85% of the total variation and PC1 contributes 26.8% and PC2 23.57% (Figure S5). The PCA for the female dataset shows slight clustering between both species and localities (Figure S4A and S4B). The clusters appear indistinct between species and localities as there are so many species / localities represented. There are distinct changes in relative eye size and position between the PCs in the female dataset (Figure S4C). In the first PC, the anterior eyes are more clustered together than the average shape. The PLEs are also vertically rotated. The second PC saw a reduction in the anterior eye sixes. The PLEs are rotated as in PC1 (Figure S4C). The male dataset PCA shows clustering for the species and locality variable (Figure S5A and S4B). The changes in eye pattern are more extreme for the male dataset than the female. Within the first PC, all eyes are significantly larger. The AMEs shift from being behind the ALEs line, to slightly in front of them (Figure S5C). In the second PC, the opposite pattern is observed, all the eyes are significantly reduced in size (Figure S5C).
The VIF analysis for the female indicated slight correlation between species and centroid size (GVIF: 2.999), and no correlation between locality and centroid size (GVIF: 1.151). There were however, certain species which had VIF values above 5, indicating a severe correlation with centroid size. These species were S. erythrognathus, S. insculptus var peddiensis and S. maraisi (Figure S6A). For the male dataset, severe correlation was found between species and centroid size (GVIF: 19.358), this is seen in the species S. erythrognathus, S. gigas, S. griswoldi, S. hewitti, S. insculptus peddiensis and Undescribed species 2. (Figure S6C). There was no correlation found between locality and centroid size for the male dataset (GVIF: 1.373).
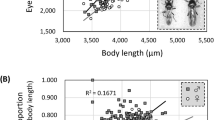
The regressions between procrustes distances and centroid size (pooled by other variables) are available in Figures S7 and S8. There is a significant relationship between shape (procrustes distances) and centroid size for both sexes (Fig. 5, Table S7). The size of an individual accounts for a large percentage of shape variability within both sexes, 7.45% for females and 13.63% for males. This percentage increases when the data are pooled by species for the female dataset (8.61%), but is not significant for males. When pooled by locality, the influence of centroid size on shape decreases, but is still significant (Table S7).
Relative eye position and size is not only significantly influenced by centroid size, but also by the species and locality / morphospecies considered (Table S8). These relationships were observed within both sexes. Pairwise tests to determine the species or locality which influence the relationship were not possible due to sample size constraints.
Clear clustering is visible in the CVA plots. The clustering is more pronounced when the data are analysed by locality/morphospecies than by species for both sexes. More of the variation in the data is accounted for in a fewer number of CVs for locality than for species. Most species show tight clustering, even though the species overlap. For the female dataset by species, there are subtle changes in ocular pattens along the first and second CV (Fig. 6A). In the locality CVA however, the changes are more pronounced, with a reduction in the AMEs and a slight rotation in the PLEs (Fig. 6B). For the male dataset, the CVA by species shows a distinct shape change (Fig. 7A). There is a reduction in the AMEs size, in the first CV the PLEs and ALEs are also reduced. There is less variation in the CVA by locality for the male dataset (Fig. 7B). In CV1 there is a reduction in AMEs, and in CV2 the ALEs are slightly larger. Cross validation could not be performed for the species variable of either dataset as some species only have one representative. For the locality variable for the female dataset the overall classification accuracy was 66.67% and for the male dataset 50.85%.
Morphospace plots of the first two CVs for the female dataset. (A) CVA by species morphospace plot. The species colours correspond to the map in Fig. 1. Shape changes in the female ocular pattern for the first and second PC are shown for all the landmarks. The original average shape is indicated in grey and the new target shape in black. (B) CVA by locality / morphospecies morphospace plot, also including the shape changes along the CVs.
Morphospace plots of the first two CVs for the male dataset. (A) CVA by species morphospace plot. The species colours correspond to the map in Fig. 1. Shape changes in the male ocular pattern for the first and second PC are shown for all the landmarks. The original average shape is indicated in grey and the new target shape in black. (B) CVA by locality / morphospecies morphospace plot, also including the shape changes along the CVs.
The between group PCA produced obvious grouping for the various datasets, but these are not as clustered as in the CVA (Figure S9 and S10). Cross validation could again not be performed for the species groupings. The overall classification accuracy for location in the female dataset was 59.64%, and for the male dataset 67.80%.
The separation of the species and morphospecies clusters are validated by the results of the procrustes ANOVA (Table S8). The ANOVA shows that there are significant relationships between shape and species groupings, as well as shape and morphospecies groupings for both sexes.
Discussion
Phylogenetic Relationship to Eye Pattern
This study is the first to test the phylogenetic signal of a trait in Stasimopus. The results show that there is significant phylogenetic signal for the relative position and size of eye patterns in the genus. The Kmult values for both sexes are above one, this shows that individuals which are more closely related will have morphologically more similar eye patterns than what would be expected under a Brownian evolutionary model (Adams, 2014b). The eye patterns of Stasimopus species can thus be used as character for classification of species (Hendrixson & Bond, 2009). The question then becomes, what may drive the observed morphological changes in eye patterns in the various species over evolutionary time? A key pattern which can be observed is that males of a species have larger principal eyes (AMEs) relative to the secondary eyes than females. The primary eyes of arachnids have higher photoreceptor sensitive and higher spatial accuracy than secondary eyes as well as having the muscularity to allow eye movements (Morehouse et al., 2017). The males may have larger AMEs due to males driving dispersal by actively seeking out females at night and thus having different visual requirements.
Females of S. rufidens and Undescribed species 1 also have larger primary eyes relative to secondary when compared to the other species. There is a fundamental gap in the scientific literature concerning the functionality of individual eye pairs within the Araneae and particularly within Mygalomorphae. This oversight is likely due to the assumption that as mygalomorphs occur in low-light environments, they must primarily rely on chemo- and mechanoreception, thus little focus being paid to vision. There is a need for this information as due to the differences in orientation, position and size of each eye pair, it is likely that they all perform different functionalities (Dacke et al., 2001; Kovoor et al., 1993). No eye pairs have never been tested in isolation for the mygalomorphs, making it difficult to draw concrete reasons as to why there may be differences in the AME of these females. One possibility may be that as the spiderlings have to leave the mothers burrow to construct their own, a larger pair of AMEs may be beneficial in more open habitats as to allow for the detection of predators through the use of active eye movements. If this is the case, then the trait of larger relative AMEs would be shared within a species by both sexes. This could not be confirmed as no male is described for S. rufidens.
A high amount of variation was found in the relative position of the PLEs. This may be a new trait to use when identifying species. These eyes would dictate the amount of peripheral vision the spider possess (Morehouse et al., 2017). The females of S. leipoldti, Undescribed species 1, Undescribed species 3 and Undescribed species 6 all have PLEs which are angled at 90 degrees, whereas the other species PLEs extend past 90 degrees towards the abdomen. This implies that these species may not require as wide a peripheral vision as the others. Possible reasons for the need of more peripheral vision may be linked to the density of vegetation around the burrow. If a burrow is not well concealed, the spider may need a wider range of vision in order to detect predators while hunting. Fine scale habitat surveys would be needed to determine if this is the case. Differences in predator and parasitoid fauna may also be a possible reason. Interestingly, these species are not sister taxa, indicating that this has evolved multiple times. The fact that mygalomorph spiders tend to be short-range endemics may explain this (Harvey, 2002; Mason et al., 2018). As genetic drift or isolated and independent selection events over time within these isolated species may have led to the convergent evolution observed.
The PMEs also appear to display variation in relative size between the species in both sexes. In S. maraisi and Undescribed species 6, both sexes have PMEs which are significantly larger than the PLEs. The same is seen for females of S. unispinosus and Undescribed species 1. Most species appear to have similar sized PMEs and PLEs, except for S. rufidens, whose female has larger PLEs than PMEs. The secondary eyes of mygalomorph spiders contain tapetum lining, which is a mechanism for doubling light paths, enabling better detection of dim light (Dahl & Granda, 1989; Morehouse et al., 2017). This has also been linked to the detection of polarized light, for which the secondary eyes are ideally situated on top of the carapace (Dacke et al., 2001). The PMEs of the Stasimopus species appear to be the most reflective of the secondary eyes, suggesting that they may be best suited for these functions. The enlargement of these eyes in some species may thus be due to different hunting times. Observational studies would be required to test this postulation. There is again, no clear phylogenetic pattern underlying this trait.
There is a significant relationship between eye pattern and body size when phylogeny is taken into account for both sexes, suggesting evolutionary allometry. This is apparent when looking at the eye pattern of larger species (e.g., S. gigas) the eye pattern is sparser and more spread across the carapace than in smaller species.
Eye Patterns as an Identifier
As clear phylogenetic signal has been confirmed for eye patterns, the next logical step is to determine to what extent it can be applied in a taxonomic context. The morphospace plots (PCA, CVA and between group PCA) show overlap between the sampled species. This is expected due to the continuous nature of the eye patterns as well as the number of species examined (Kallal et al., 2022). Despite this, species still form discrete groupings, implying that the differences between species is greater than within species, this is further supported by the results of the procrustes ANOVA. It is likely that some of the overlap could be reduced by increasing the sample size of each species, as many are only represented by one specimen. The apparent shape differences between the PCs and CVs show that there are distinct shape differences between the various species. This is demonstrated clearly when looking at the average thin plate splines if the sequenced species. Each species has a distinct eye pattern, but a larger sample size would be necessary to create average shapes for each species.
Another important aspect is that some synoptic species appear to be separate in morphospace (e.g., S. maraisi and Undescribed species 6). This could be a useful way of distinguishing species which co-occur in a quantitative manner. There are very few characters which are reliably used to distinguish species not only in Stasimopus but the Mygalomorphae generally (Hendrixson & Bond, 2009). Thus, finding a continuous character that can be used to do so would aid in this greatly. This does however, indicate that the morphospecies groupings based on geographic proximity and similarity may be problematic and need to be revised. Cross validation of the locality variable shows fair results. Accuracy is between 50 and 70% for both sexes, which is surprising given the small dataset and geographic groupings.
The results of the regression analyses showed a significant correlation between shape and centroid size for both sexes. These larger specimens are also the data points that affected the VIF results as shape and size are linked. As shown, evolutionary allometry is present in the genus, thus the variation accounted for by this relationship is important when attempting to determine species classifications and should not be removed in subsequent studies. When this relationship is pooled by species, it accounts for below 9% of the variation and is thus, although important, is not a main factor driving shape variation among species. The relationship of size and shape in the eye patterns of Stasimopus is important to note as it contributes to shape in larger specimens, but it is not a main contributor to overall eye shape in the genus.
The significant relationship between both species and locality with shape in both sexes reiterates that the relative eye position and size can be used as an identifier. It would be useful to conduct pairwise analyses to determine which species can be differentiated by this technique, this would be a good avenue for future research if a larger sample size can be obtained.
There is clear variation in the relative positions and sizes of the eye patterns in both the males and females of the genus. These differences have qualitatively been used to describe and identify species historically (Engelbrecht & Prendini, 2011; Hendrixson & Bond, 2004; Hewitt, 1910; Pocock, 1902). This study provides the first statistical evidence that this trait carries phylogenetic signal and should and actually can be used for this purpose. We show the power of this method to elucidate the evolutionary history and adaptations of the spider genus. We also show that using geometric morphometrics to quantitatively differentiate morphospecies using this character is possible and there is potential for the methodology to be applied at species level, but this requires validation with more samples or additional species.
References
Adams, D. C. (2014a). A method for assessing phylogenetic least squares models for shape and other high-dimensional multivariate data. Evolution, 68(9), 2675–2688. https://doi.org/10.1111/evo.12463.
Adams, D. C. (2014b). A generalized K statistic for estimating phylogenetic Signal from shape and other high-dimensional Multivariate Data. Systematic Biology, 63(5), 685–697. https://doi.org/10.1093/sysbio/syu030.
Adams, D. C., Collyer, M. L., Kaliontzopoulou, A., & Baken, E. K. (2021). Geomorph: Software for geometric morphometric analyses (4.0). R package.
Ayres, D. L., Darling, A., Zwickl, D. J., Beerli, P., Holder, M. T., Lewis, P. O., Huelsenbeck, J. P., Ronquist, F., Swofford, D. L., Cummings, M. P., Rambaut, A., & Suchard, M. A. (2012). BEAGLE: An application Programming Interface and High-Performance Computing Library for statistical phylogenetics. Systematic Biology, 61(1), 170–173. https://doi.org/10.1093/sysbio/syr100.
Baken, E. K., Collyer, M. L., Kaliontzopoulou, A., & Adams, D. C. (2021). Geomorph v4.0 and gmShiny: Enhanced analytics and a new graphical interface for a comprehensive morphometric experience. Methods in Ecology and Evolution, 12(12), 2355–2363. https://doi.org/10.1111/2041-210X.13723.
Bakkes, D. K. (2017). Evaluation of measurement error in rotational mounting of larval Rhipicephalus (Acari: Ixodida: Ixodidae) species in geometric morphometrics. Zoomorphology, 136(3), 403–410. https://doi.org/10.1007/s00435-017-0357-8.
Bond, J. E., & Beamer, D. A. (2006). A morphometric analysis of mygalomorph spider carapace shape and its efficacy as a phylogenetic character (Araneae). Invertebrate Systematics, 20(1), 1–7. https://doi.org/10.1071/IS05041.
Bond, J. E., Hendrixson, B. E., Hamilton, C. A., & Hedin, M. (2012). A reconsideration of the classification of the spider infraorder Mygalomorphae (Arachnida: Araneae) based on three nuclear genes and morphology. Plos One, 7(6), e38753. https://doi.org/10.1371/journal.pone.0038753.
Clavel, J., Escarguel, G., & Merceron, G. (2015). mvMORPH: An R package for fitting multivariate evolutionary models to morphometric data. Methods in Ecology and Evolution, 6(11), 1311–1319. https://doi.org/10.1111/2041-210X.12420.
Clavel, J., Aristide, L., & Morlon, H. (2019). A penalized Likelihood Framework for high-dimensional phylogenetic comparative methods and an application to New-World Monkeys Brain Evolution. Systematic Biology, 68(1), 93–116. https://doi.org/10.1093/sysbio/syy045.
Collyer, M. L., & Adams, D. C. (2018). RRPP: An R package for fitting linear models to high-dimensional data using residual randomization. Methods in Ecology and Evolution, 9(7), 1772–1779. https://doi.org/10.1111/2041-210X.13029.
Collyer, M. L., & Adams, D. C. (2021). RRPP: Linear Model evaluation with Randomized Residuals in a permutation Procedure (4(vol.). R package. 0.
R Core Team (2017). R: A language and environment for statistical computing (3.3.3). R Foundation for Statistical Computing. www.R-project.org
Rambaut, A. (2016). FigTree (1.4.3). Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/.
Costa-schmidt, L. E., & de Araujo, A. M. (2010). Genitalic variation and taxonomic discrimination in the semi-aquatic spider genus Paratrechalea (Araneae: Trechaleidae). The Journal of Arachnology, 38, 242–249. https://doi.org/10.1636/JOA_A09-75.1.
Crews, S. C. (2011). A revision of the spider genus Selenops Latreille, 1819 (Arachnida, Araneae, Selenopidae) in North America, Central America and the Caribbean. Zookeys, 105, 1–182. https://doi.org/10.3897/zookeys.105.724.
Dacke, M., Doan, T. A., & O’Carroll, D. C. (2001). Polarized light detection in spiders. Journal of Experimental Biology, 204(14), 2481–2490. https://doi.org/10.1242/jeb.204.14.2481.
Dahl, R. D., & Granda, A. M. (1989). Spectral sensitivities of photoreceptor in the ocelli of the tarantula, Aphonopelma chalcodes (Araneae, Theraphosidae). Journal of Arachnology, 17, 195–205.
Doyle, J. J. (1992). Gene trees and species trees: Molecular systematic one-character taxonomy. Systematic Botany, 17, 144–163. https://doi.org/10.2307/2419070.
Drummond, A. J., Ho, S. Y. W., Phillips, M. J., & Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PLoS Biology, 4(5), 0699–0710. https://doi.org/10.1371/journal.pbio.0040088.
Dunlop, J. A., Penney, D., & Jekel, D. (2020). A summary list of fossil spiders and their relatives. World Spider Catalog; Natural History Museum Bern. http://wsc.nmbe.ch.
Engelbrecht, I., & Prendini, L. (2011). Assessing the taxonomic resolution of southern african trapdoor spiders (Araneae: Ctenizidae; Cyrtaucheniidae; Idiopidae) and implications for their conservation. Biodiversity and Conservation, 20(13), 3101–3116. https://doi.org/10.1007/s10531-011-0115-z.
Engelbrecht, I., & Prendini, L. (2012). Cryptic diversity of south african trapdoor spiders: Three new species of Stasimopus Simon, 1892 (Mygalomorphae, Ctenizidae), and redescription of Stasimopus robertsi Hewitt, 1910. American Museum Novitates, 3732, 1–42. https://doi.org/10.1206/3732.2.
Fernández-Montraveta, C., & Marugán-Lobón, J. (2017). Geometric morphometrics reveals sex-differential shape allometry in a spider. PeerJ, 5, e3617. https://doi.org/10.7717/peerj.3617.
Forthofer, R. N., Lee, E. S., & Hernandez, M. (2007). Linear Regression. Biostatistics, 349–386. https://doi.org/10.1016/B978-0-12-369492-8.50018-2.
Fox, J., & Weisberg, S. (2019). An R companion to Applied Regression. Third). Sage.
Fruciano, C. (2016). Measurement error in geometric morphometrics. Development Genes and Evolution, 226(3), 139–158. https://doi.org/10.1007/s00427-016-0537-4.
Goodall, C. (1991). Procrustes Methods in the statistical analysis of shape. Journal of the Royal Statistical Society: Series B (Methodological), 53(2), 285–321. https://doi.org/10.1111/j.2517-6161.1991.tb01825.x.
Harvey, M. S. (2002). Short-range endemism among the australian fauna: Some examples from non-marine environments. Invertebrate Systematics, 16, 555–570. https://doi.org/10.1071/IS02009.
Hendrixson, B. E., & Bond, J. E. (2004). A new species of Stasimopus from the Eastern Cape Province of South Africa (Araneae, Mygalomorphae, Ctenizidae), with notes on its natural history. Zootaxa, 619, 1–14. https://doi.org/10.5281/zenodo.10101.
Hendrixson, B. E., & Bond, J. E. (2009). Evaluating the efficacy of continuous quantitative characters for reconstructing the phylogeny of a morphologically homogeneous spider taxon (Araneae, Mygalomorphae, Antrodiaetidae, Antrodiaetus). Molecular Phylogenetics and Evolution, 53(1), 300–313. https://doi.org/10.1016/j.ympev.2009.06.001.
Hewitt, J. (1910). Description of two trapdoor spiders from Pretoria (female of Acanthodon pretoriae Poc and Stasimopus robertsi, n. sp). Annals of the Transvaal Museum, 2, 74–76.
Jocque, R., & Dippenaar-Schoeman, A. S. (2007). Spider families of the world (Second ed.). Royal Museum for Central Africa.
Kallal, R. J., de Miranda, G. S., Garcia, E. L., & Wood, H. M. (2022). Patterns in schizomid flagellum shape from elliptical Fourier analysis. Scientific Reports, 12(1), 3896. https://doi.org/10.1038/s41598-022-07823-y.
Katoh, K. (2005). MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Research, 33, 511–518. https://doi.org/10.1093/nar/gki198.
Katoh, K., & Toh, H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics, 9, 286–298. https://doi.org/10.1093/bib/bbn013.
Klingenberg, C. P. (2011). MorphoJ: An integrated software package for geometric morphometrics. Molecular Ecology Resources, 11, 353–357. https://doi.org/10.1111/j.1755-0998.2010.02924.x.
Klingenberg, C. P. (2016). Size, shape, and form: Concepts of allometry in geometric morphometrics. Development Genes and Evolution, 226(3), 113–137. https://doi.org/10.1007/s00427-016-0539-2.
Klingenberg, C. P., & Gidaszewski, N. A. (2010). Testing and quantifying phylogenetic signals and homoplasy in morphometric data. Systematic Biology, 59(3), 245–261. https://doi.org/10.1093/sysbio/syp106.
Kovoor, J., Cuevas, A. M., & Escobar, J. O. (1993). Microanatomy of the anterior median eyes and its possible relation to polarized-light reception in lycosa tarentula (Araneae, lycosidae). Bolletino Di Zoologia, 60(4), 367–375. https://doi.org/10.1080/11250009309355841.
Kück, P., & Meusemann, K. (2010). FASconCAT: Convenient handling of data matrices. Molecular Phylogenetics and Evolution, 56, 1115–1118. https://doi.org/10.1016/j.ympev.2010.04.024.
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution, 35(6), 1547–1549. https://doi.org/10.1093/molbev/msy096.
Maddison, W. P. (1997). Gene trees in species trees. Systematic Bibliology, 46, 523–536. https://doi.org/10.1093/sysbio/46.3.523.
Maindonald, J. H., & Braun, W. J. (2020). DAAG: Data Analysis and Graphics Data and Functions (1(vol.). R package. 24.
Manthey, L., & Ousley, S. D. (2020). Geometric morphometrics. Statistics and Probability in Forensic Anthropology, 289–298. https://doi.org/10.1016/B978-0-12-815764-0.00023-X.
Mason, L. D., Wardell-Johnson, G., & Main, B. Y. (2018). The longest-lived spider: Mygalomorphs dig deep, and persevere. Pacific Conservation Biology, 24(2), 203. https://doi.org/10.1071/PC18015.
McKeown, A. H., & Schmidt, R. W. (2013). Geometric Morphometrics. In DiGangi, E.A & Moore, M. K. (Eds.), Research Methods in Human Skeletal Biology (pp. 325–359). Elsevier. https://doi.org/10.1016/B978-0-12-385189-5.00012-1.
Michonneau, F. (2017). Using GMYC for species delimitation. Zenodo. https://doi.org/10.5281/zenodo.838259.
Mitteroecker, P., & Gunz, P. (2009). Advances in geometric morphometrics. Evolutionary Biology, 36, 235–247. https://doi.org/10.1007/s11692-009-9055-x.
Morehouse, N. I., Buschbeck, E. K., Zurek, D. B., Steck, M., & Porter, M. L. (2017). Molecular evolution of spider vision: New opportunities, familiar players. Biological Bulletin, 233(1), 21–38. https://doi.org/10.1086/693977.
Nakagawa, S., Kar, F., O’Dea, R. E., Pick, J. L., & Lagisz, M. (2017). Divide and conquer? Size adjustment with allometry and intermediate outcomes. BMC Biology, 15(1), 107. https://doi.org/10.1186/s12915-017-0448-5.
Opatova, V., Hamilton, C. A., Hedin, M., de Oca, L. M., Král, J., & Bond, J. E. (2020). Phylogenetic systematics and evolution of the Spider Infraorder Mygalomorphae using genomic Scale Data. Systematic Biology, 69(4), 671–707. https://doi.org/10.1093/sysbio/syz064.
Pirouz, D. M. (2006). An overview of partial least squares. SSRN Electronic Journal. https://doi.org/10.2139/ssrn.1631359.
Pocock, R. I. (1902). Descriptions of some new species of African Solifugae and Araneae. Annals and Magazine of Natural History, 7(10), 6–27.
Polotow, D., & Brescovit, A. D. (2014). Phylogenetic analysis of the tropical wolf spider subfamily Cteninae (Arachnida, Araneae, Ctenidae). Journal of the Linnean Society, 170, 333–361. https://doi.org/10.1111/zoj.12101.
Posada, D. (2008). jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution, 25(7), 1253–1256. https://doi.org/10.1093/molbev/msn083.
Prenter, J., Montgomery, I. W., & Elwood, R. W. (1995). Multivariate morphometrics and sexual dimorphism in the orb-web spider Metellina segmentata (Clerck, 1757) (Araneae, Metidae). Biological Journal of the Linnean Society, 55, 345–354.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., & Suchard, M. A. (2018). Posterior summarization in bayesian phylogenetics using Tracer 1.7. Systematic Biology, 67(5), 901–904. https://doi.org/10.1093/sysbio/syy032.
Ringle, C. M., Sarstedt, M., Mitchell, R., & Gudergan, S. P. (2018). Partial least squares structural equation modelling in HRM research. The International Journal of Human Resource Management, 31(12), 1617–1643. https://doi.org/10.1080/09585192.2017.1416655.
Rodriguez-Mendoza, R. (2013). Population structure of the bluemouth, Helicolenus dactylopterus (Teleostei: Sebastidae), in the Northeast Atlantic and Mediterranean using geometric morphometric techniques [Thesis]. University of Vigo.
Rohlf, F. J. (2015). The tps series of software. Hystrix the Italian Journal of Mammalogy, 26, 1–4. https://doi.org/10.4404/hystrix-26.1-11264.
Rohlf, F. J., & Corti, M. (2000). Use of two-block partial least-squares to study covariation in shape. Systematic Biology, 49(4), 740–753. https://doi.org/10.1080/106351500750049806.
Rohlf, F. J., & Marcus, L. F. (1993). A revolution in Morphometrics. TREE, 8(4), 129–132. https://doi.org/10.1016/0169-5347(93)90024-J.
Schlager, S. (2013). Soft-tissue reconstruction of the human nose: population differences and sexual dimorphism [Thesis]. Freiburg University.
Seetah, T. K. (2014). Geometric morphometrics and environmental archaeology. In C. Smith (Ed.), Encyclopedia of global archaeology. Springer. https://doi.org/10.1007/978/1-4419-0465-2.
Selden, P. A. (2002). First british mesozoic spider, from cretaceous amber of the Isle of Wight, southern England. Palaeontology, 45, 973–983. https://doi.org/10.1111/1475-4983.00271.
Selden, P. A., & Gall, J. C. (1992). A triassic mygalomorph spider from the northern Vosges, France. Palaeontology, 35(1), 211–235.
Simon, E. (1892). Histoire naturelle des araignées. Deuxième Édition Tome Premier, 1–256.
Spasojevic, T., Kropf, C., Nentwig, W., & Lasut, L. (2016). Combining morphology, DNA sequences and morphometrics: Revising closely related species in the orb-weaving spider genus Araniella (Araneae, Araneidae). Zootaxa, 4111(4), 488–470. https://doi.org/10.11646/zootaxa.4111.4.6.
Suchard, M. A., Lemey, P., Baele, G., Ayres, D. L., Drummond, A. J., & Rambaut, A. (2018). Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evolution, 4(1), vey016. https://doi.org/10.1093/ve/vey016.
World Spider Catalog (2022). World Spider Catalog Version 23.5. Natural History Museum Bern. Retrieved June 5, 2022 from https://doi.org/10.24436/2.
Wunderlich, J. (1998). Beschreibung der ersten fossilen Spinnen der Unterfamilien Mysmeninae (Anapidae) und Erigoninae (Linyphiidae) im Dominikanischen Bernstein (Arachnida: Araneae). Entomologische Zeitschrift, 108, 363–367.
Wunderlich, J. (2000). Zwei neue Arten der Familie Falltürspinnen (Araneae: Ctenizidae) aus dem Baltischen Bernstein. Entomologische Zeitschrift, 110, 345–348.
Zelditch, M. L., Swiderski, D. L., Sheets, H. D., & Fink, W. L. (2004a). Landmarks. In M. L. Zelditch, D. L. Swiderski, & H. D. Sheets (Eds.), Geometric morphometrics for biologists: A primer (2nd ed., pp. 23–50). Elsevier Academic Press.
Zelditch, M. L., Swiderski, D. L., Sheets, H. D., & Fink, W. L. (2004b). Superimposition methods. In M. L. Zelditch, D. L. Swiderski, & H. D. Sheets (Eds.), Geometric morphometrics for biologists: A primer (2nd ed., pp. 105–128). Elsevier Academic Press.
Zelditch, M. L., Swiderski, D. L., Sheets, H. D., & Fink, W. L. (2004c). Theory of shape. In M. L. Zelditch, D. L. Swiderski, & H. D. Sheets (Eds.), Geometric morphometrics for biologists: A primer (2nd ed., pp. 73–104). Elsevier Academic Press.
Žikić, V., Stanković, S. S., Petrović, A., Ilić Milošević, M., Tomanović, Ž., Klingenberg, C. P., & Ivanović, A. (2017). Evolutionary relationships of wing venation and wing size and shape in Aphidiinae (Hymenoptera: Braconidae). Organisms Diversity & Evolution, 17(3), 607–617. https://doi.org/10.1007/s13127-017-0338-2.
Acknowledgements
We would like to acknowledge Tshepiso Majelantle and Elmé Brand for their assistance on fieldwork for sample collection. We thank Carmelo Fruciano for his advice with framing the research question for this manuscript. Equipment for field work was supplied by the Agricultural Research Council. We would like to thank the following institutions for loaning the specimens required for specimen identification: Albany Museum, Grahamstown, South Africa; The National Museum, Bloemfontein, South Africa; Ditsong Museum of Natural History, Pretoria, South Africa; Iziko Museum of Cape Town, South Africa; Museum für Naturkunde, Berlin, Germany; National Collection of Arachnida, Pretoria, South Africa.
Funding
The National Research Foundation (NRF) are acknowledged for funding to the second author. The first author would also like to acknowledge the National Research Foundation for funding (Grant no: 120190 and 139125). This work is based on the research supported in part by the Karoo BioGaps Project (NRF Grant Number 98864), which was awarded through the Foundational Biodiversity Information Programme (FBIP), a joint initiative of the Department of Science Innovation (DSI), the National Research Foundation (NRF) and the South African National Biodiversity Institute (SANBI).
Open access funding provided by University of Pretoria.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation was performed by Shannon Brandt. Data collection was performed by Shannon Brandt, Catherine Sole and Robin Lyle. Analyses were performed by Shannon Brandt and assisted by Christian Pirk. The first draft of the manuscript was written by Shannon Brandt and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
No conflicting interests to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Brandt, S., Sole, C., Lyle, R. et al. Geometric Morphometric Analysis of Ocular Patterns as a Species Identifier in the South African Endemic Trapdoor Spider Genus Stasimopus Simon, 1892 (Araneae, Mygalomorphae, Stasimopidae). Evol Biol 50, 350–364 (2023). https://doi.org/10.1007/s11692-023-09609-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11692-023-09609-0