Abstract
Many trait measurements are size-dependent, and while we often divide these traits by size before fitting statistical models to control for the effect of size, this approach does not account for allometry and the intermediate outcome problem. We describe these problems and outline potential solutions.
Similar content being viewed by others
Adjusting for size in statistical analysis
Biologists measure traits of organisms, characterizing a range of features including morphology, physiology and behaviour. Many of these traits are size-dependent. For example, larger animals eat more and larger plants absorb more than smaller counterparts. However, when size is not our trait of interest, we often want to know values of focal traits after controlling for the effect of size. An intuitive way to account for organismal size is to divide a trait of interest by size (for example, the amount of food consumed or nutrient absorbed divided by mass or length). This method (hereafter called the “division” method) generates size-adjusted trait values, which can be used for statistical analyses. Indeed, the use of the division method is prevalent in the literature [1,2,3,4], but is it correct? The division method poses two major problems because, first, we assume a linear relationship between adjusted traits and size and, second, an experimental treatment often affects not only a trait of interest but also other variables, such as size. Here, we deliberate on these two problems and provide potential solutions.
Allometric scaling of organismal traits
The power law (meaning non-linear) relationship between body size and body parts is believed to have been first described by the evolutionary biologist Julian Huxley, (a grandson of “Darwin’s bulldog” Thomas Huxley) on the basis of his study of claw size and body size in fiddler crabs [5]. This non-linear relationship was later termed “allometry” (meaning “different measure”) [6]. Huxley’s original study was on ontogenetic allometry—the relationship between two traits while an organism is growing [7]. This type of allometry is distinguished from the two other types: evolutionary allometry and static allometry [7]. Evolutionary allometry concerns between-species variation in the relationship between traits (see [8] for an in-depth review), whereas static allometry concerns the relationship between two traits of mature individuals from the same species. Here, we focus mainly on static allometry.
Historically, allometric studies have focused on morphological traits. However, physiological traits (such as cellular and drug metabolism [9, 10]) and behavioural traits (for instance, food consumption [2, 11]) also scale allometrically with organismal size. Traits follow the power law relationship described by Huxley [5], which is expressed as:
where y [trait] is the focal variable (trait), x [size] is the size variable, and a and b are constants (parameters) estimated from data (Fig. 1a). In order to linearize this non-linear equation, we can take the natural logarithm of both sides; thus, we get:
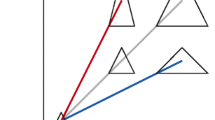
Conceptual plots for allometric relationships. a Three different types of allometric relationships between food intake (a focal trait; on the y axis) and size (on the x axis) with different exponents, b, and a fixed slope, a (Eq. 1); note that when b = 1, the relationship is linear. b When b is close to 1 (b = 0.9), the relationship becomes nearly linear without log-transformation (dotted line). c, d Even when b is not close to 1 (b = 0.5), whether the relationship is non-linear depends on how the data are distributed; the non-linear relationship in d could be much better approximated by a linear line than that in c. e, f Notably, the same slopes (b = 0.5) can be estimated as having different slopes if not log-transformed due to having different values for a, as in e, or being on different parts of a non-linear curve, as in f
where y [ln(trait)] is ln(y [trait]) and x [ln(size)] is ln(x [size]), and ln is the log e transformation (see [12, 13] for discussions of allometry and the log transformation).
Now consider an experimental study that compares a size-dependent trait of two groups (for example, control vs. treatment; note that non-experimental groups—such as males vs. females, or two natural populations—could be also used). When we use the division method to adjust for size in the analysis, we use the following linear model:
where y [trait/size] is a variable derived from the focal trait divided by size, x [group] is a “dummy variable”, which takes the value 0 or 1 to indicate the presence or absence of a particular effect in order to sort data in mutually exclusive groups (for instance, 0 = control and 1 = experiment), b 0 is the intercept, b 1 is the slope (or in this case, the difference between the control and experimental group: experimental or treatment effect), e is the residual (error) term (which represents deviations from the regression line), and the subscript i indicates the i th value (i = 1…n, n = sample size; this linear model, Eq. 3, is equivalent to a t-test comparing the two groups). However, this model is not ideal because the division method (Eq. 3) creates a ratio variable (y [trait/size]), the distributional properties of which may not meet an important assumption of a linear model: the residuals are normally distributed [1]. Furthermore, we are not able to estimate the allometric scaling exponent b.
We can improve Eq. 3 by fitting size as a predictor variable because it avoids creating a ratio variable in the response variable. Then, we have:
where b 2 is the slope for size, and the other symbols are as above. This approach is again common in the biological literature. However, both Eq. 3 (the division method) and Eq. 4 assume a linear relationship between a trait and size; they ignore the allometric relationship shown in Eqs. 1 and 2. Therefore, a better statistical model would be to linearize the equation by taking natural logs:
where b 0 and b 2 correspond to lna and b in Eq. 2, respectively, and the other symbols are as above. It is also notable that Eqs. 4 and 5 could produce comparable results, depending on the exponent of the power law relationship, b, and the distribution of trait data (Fig. 1b–d). However, Eq. 4, like Eq. 3, could lead not only to a spurious treatment effect, b 1 [2], but also to spurious interactions (which means that the control and experimental groups have different slopes, b 2 as described in Fig. 1e, f); see the next section for modeling the interaction (that is, different slopes). However, it turns out that even Eq. 5 can provide an incorrect estimate of b 1.
Experimental treatments and intermediate outcomes
An experimental treatment is intended to change a focal variable, but it often affects other unintended variables, referred to as mediators or intermediate outcomes [14]. For example, maternal dietary conditions, such as dietary restriction or over-nutrition, may influence offspring size (x), as well as offspring food intake (the focal trait; y) [2, 11]. If we know that offspring body mass and offspring food intake are correlated, we may want to account for the effect of offspring size when assessing experimental effects on offspring food intake. However, as both offspring body mass and food intake are measured after the treatment has been applied (offspring body mass cannot be measured before maternal diet is manipulated), we do not know the chain of causation. Imagine two scenarios. In scenario A, offspring body mass and food intake are mechanistically linked, and the maternal diet treatment subsequently affects both traits (direct effects; Fig. 2a). Alternatively, in scenario B, the treatment affects offspring body size (direct effect), which then influences offspring food intake (indirect effect), as well as a potential direct effect of the treatment on offspring food intake (Fig. 2b).
Two scenarios of the relationship among an experimental treatment, a trait of interest (focal variable, y) and an intermediate outcome (x). a The treatment affects both x and y, and therefore x and y are correlated (dotted line with a double-headed arrow) but x does not affect y. b The treatment affects both x and y, and then x also affects y
In both scenarios, we would observe a treatment effect in both x and y, but when we correct for x (as in Eq. 5), the direct effect of the treatment on y could diminish, disappear or even reverse (known as Lord’s paradox) [15]. If scenario A is true, then correcting for x leads to an underestimation of the direct treatment effect (b 1 in Eq. 5) on y (known as over-adjustment bias) [14, 16]. If scenario B is true, then not correcting for x leads to an overestimation of the direct effect on y (specifically, direct effects plus indirect effects). Unfortunately, we are unlikely to disentangle the two scenarios unless we have prior knowledge of the mechanistic underpinnings of these relationships. Furthermore, even if we know that scenario B is true, we must assume that all subjects’ size responds in the same way to the treatment (for example, every subject gains 200 g) to obtain the correct b 1 (the direct effect) using Eq. 5, although this seems unlikely. Therefore, it has been suggested that we should not add an intermediate outcome (such as size) to the model regardless of the scenarios [14], as follows:
where the symbols are as above. Then, at least we will get the total (direct and indirect) effect on y as the experimental effect, b 1 (for further discussion of intermediate outcomes and their problems see [14,17,, 16–18]).
Importantly for our discussion here, this solution means that we are unable to estimate the allometric scaling component. Here we suggest a workaround using a linear model [19] (see reference [20] for an example), which can be written as:
where the subscript “wgc” stands for within-group centering, which adjusts values (ln(size)) for two (or more) different groups separately by setting respective group means as zero (Fig. 3). Figure 3a visualizes what such centering does to a size variable; the order of the two transformations for x [wgc(ln(size))] (log first, then center) is particularly important, as one cannot take the logarithm of negative values. The within-group centering separates the experimental effect on size (indirect effect), so that the indirect effect is now absorbed into b 1. With this approach, we can model an allometric relationship, although we cannot obtain a “size-corrected” experimental effect (an unbiased direct effect, but see [21, 22] for a potential issue and solution; Fig. 2). Note that the estimates of b 1 from Eqs. 6 and 7 are the same, as are the estimates of b 2 (that is, the allometric scaling exponent) from Eqs. 5 and 7.
Another way to do such adjustment is through z-transformation instead of centering, which scales distributions of ln(size) for both the control and experimental group to have the mean of 0 and standard deviation of 1. These two methods (centering and z-transformation) are equivalent when the variances of the two groups are the same.
However, if slopes differ between the two groups after z-transformation, it should be checked whether the transformation caused the significant differences, which may happen if the variances for ln(size) to differ between the two groups. (Figure 3b; see below for how to detect differences in slopes). The choice of transformation to use (z-transformation or centering) should not affect the experimental effect, but z-transformation could lead to confusion if one is interested in obtaining allometric parameters (a and b in Eqs. 1 and 2). Centering size on the log scale may also be easier to interpret than z-transformation (the issue of different variances described above aside). However, z-transformation has some advantages over centering when applied normally to a predictor variable (meaning, not within-group transformations). For example, z-transformation can help another aspect of interpretation because regression coefficients of continuous variables become comparable (in that they are standardized beta coefficients) [23].
Notably, we still assume that the variances for the response variable are homogenous in this model (in fact, this is true for all the models above). When this assumption is not met (that is, the response variable, y, is heteroscedastic), either we can model different variances in the response between two groups, or we can use “robust” statistical estimators, which take heteroscedasticity into account (for details on how, see [24]). We suspect that when variance in body sizes between two groups is different, it is likely that a trait of interest is heteroscedastic.
Equations 5 and 7 only model a change in the intercepts between two groups, which corresponds to a change in ln(a) in Eq. 2 (Fig. 3a). It is possible that some experimental treatments could affect the slope, which corresponds to a change in the exponent b in Eqs. 1 and 2. Biologically, this parameter, b, is less likely to change than a, at least for some allometric relationships (for an example see reference [10]). Nonetheless, we should probably check for such a change. An equation that models different slopes between two groups can be written as:
where b 3 is the difference in slopes between the control and experimental groups, and x [group] x [wgc(ln(size))] is an interaction term between x [group] and x [wgc(ln(size))] (Figs. 1 and 3). An example of implementing the above procedures in the statistical environment R [25] is provided in the Additional file 1. In this supplement, we refer to the assumption that size is measured without error in linear models such as Eqs. 4, 5, 7 and 8 [26, 27] and also provide a solution to this problem when this assumption is not met.
Divide and conquer? No, leave them alone!
We have described the two major shortcomings of the division method, so far focusing on scenarios when we use the ratio variable (for instance, dividing a focal trait by size) as a response variable (y). However, it is just as common to find the ratio variable being used as a predictor variable (x). Among the many issues of using ratios as predictors, there is one problem that is very general and straightforward to describe [1]. When we have two variables (or traits), A and B, their ratio is A/B. The variable A/B fitted as a predictor can be considered an interaction term, because A/B can be re-expressed as AB−1 (compare it with x [group] x [wgc(ln(size))] in Eq. 8). Therefore, we should also fit A and B−1 as predictors (main effects), along with the interaction term (AB−1) [1]. More generally, it is usually not advisable to create and fit a derived variable (in other words, a variable comprised of more than one variable, such as, A/B, AB2) to a model. For linear modelling, raw measurements or their direct transformations should be used to control for confounding effects. Finally, because the division method and other inappropriate modelling procedures could lead to spuriously significant results and/or biased effect size estimates [1, 2], correct modelling practice is essential to avoid exacerbating the current “reproducibility crisis” [28, 29].
References
Kronmal RA. Spurious correlation and the fallacy of the ratio standard revisited. J R Stat Soc a Stat. 1993;156(3):379–92.
Lagisz M, Blair H, Kenyon P, Uller T, Raubenheimer D, Nakagawa S. Transgenerational effects of caloric restriction on appetite: a meta-analysis. Obes Rev. 2014;15(4):294–309.
Raubenheimer D. Problems with ratio analysis in nutritional studies. Funct Ecol. 1995;9(1):21–9.
Sollberger S, Ehlert U. How to use and interpret hormone ratios. Psychoneuroendocrinology. 2016;63:385–97.
Huxley JS. Constant differential growth ratios and their significance. Nature. 1924;114:895–6.
Huxley JS, Tessier G. Terminology of relative growth. Nature. 1936;137:780–1.
Cheverud JM. Relationships among ontogenetic, static, and evolutionary allometry. Am J Phys Anthropol. 1982;59(2):139–49.
Shingleton AW. Allometry: the study of biological scaling. Nat Educ Knowl. 2010;3(10):2.
Caldwell GW, Masucci JA, Yan ZY, Hageman W. Allometric scaling of pharmacokinetic parameters in drug discovery: can human CL, VSS and t1/2 be predicted from in-vivo rat data? Eur J Drug Metab Pharmacokinet. 2004;29(2):133–43.
White CR, Seymour RS. Allometric scaling of mammalian metabolism. J Exp Biol. 2005;208(9):1611–9.
Lagisz M, Blair H, Kenyon P, Uller T, Raubenheimer D, Nakagawa S. Little appetite for obesity: meta-analysis of the effects of maternal obesogenic diets on offspring food intake and body mass in rodents. Int J Obes. 2015;39(12):1669–78.
Mascaro J, Litton CM, Hughes RF, Uowolo A, Schnitzer SA. Is logarithmic transformation necessary in allometry? Ten, one-hundred, one-thousand-times yes. Biol J Linn Soc. 2014;111(1):230–3.
Packard GC. Is logarithmic transformation necessary in allometry? Biol J Linn Soc. 2013;109(2):476–86.
Gelman A, Hill J. Data analysis using regression and multilevel/hierarchical models. Cambridge: Cambridge University Press; 2006.
Tu YK, Gunnell D, Gilthorpe MS. Simpson's paradox, Lord's paradox, and suppression effects are the same phenomenon--the reversal paradox. Emerg Themes Epidemiol. 2008;5:2.
Schisterman EF, Cole SR, Platt RW. Overadjustment bias and unnecessary adjustment in epidemiologic studies. Epidemiology. 2009;20(4):488–95.
Bulterys M, Morgenstern H. Confounding or intermediate effect? An appraisal of iatrogenic bias in perinatal AIDS research. Paediatr Perinat Epidemiol. 1993;7(4):387–94.
Vansteelandt S, Bekaert M, Claeskens G. On model selection and model misspecification in causal inference. Stat Methods Med Res. 2012;21(1):7–30.
Schneider BA, Avivi-Reich M, Mozuraitis M. A cautionary note on the use of the Analysis of Covariance (ANCOVA) in classification designs with and without within-subject factors. Front Psychol. 2015;6:474.
Crean AJ, Kopps AM, Bonduriansky R. Revisiting telegony: offspring inherit an acquired characteristic of their mother's previous mate. Ecol Lett. 2014;17(12):1545–52.
Ludtke O, Marsh HW, Robitzsch A, Trautwein U, Asparouhov T, Muthen B. The multilevel latent covariate model: A new, more reliable approach to group-level effects in contextual studies. Psychol Methods. 2008;13(3):203–29.
Phillimore AB, Hadfield JD, Jones OR, Smithers RJ. Differences in spawning date between populations of common frog reveal local adaptation. Proc Natl Acad Sci U S A. 2010;107(18):8292–7.
Schielzeth H. Simple means to improve the interpretability of regression coefficients. Methods Ecol Evol. 2010;1(2):103–13.
Cleasby IR, Nakagawa S. Neglected biological patterns in the residuals A behavioural ecologist's guide to co-operating with heteroscedasticity. Behav Ecol Sociobiol. 2011;65(12):2361–72.
R Development Core Team. R. A language and environment for statistical computing. 340th ed. Vienna: R Foundation for Statistical Computing; 2017.
Forstmeier W. Women have relatively larger brains than men: a comment on the misuse of general linear models in the study of sexual dimorphism. Anat Rec. 2011;294(11):1856–63.
Kilmer JT, Rodriguez RL. Ordinary least squares regression is indicated for studies of allometry. J Evol Biol. 2017;30(1):4–12.
Aarts AA, Anderson JE, Anderson CJ, Attridge PR, Attwood A, Axt J, et al. Estimating the reproducibility of psychological science. Science. 2015;349:aac4716.
Nakagawa S, Parker TH. Replicating research in ecology and evolution: feasibility, incentives, and the cost-benefit conundrum. BMC Biol. 2015;13:88.
Acknowledgements
The authors thank Dan Noble, Russell Bonduriansky, and Holger Schielzeth for comments and discussions. SN is supported by an ARC Future Fellowship (FT130100268) and JLP by a SNSF Early Postdoc.Mobility fellowship (P2ZHP3_164962).
Author information
Authors and Affiliations
Contributions
SN and ML conceived the idea. All authors contributed to developing the idea further and writing of the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Supplementary material. (PDF 320 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Nakagawa, S., Kar, F., O’Dea, R.E. et al. Divide and conquer? Size adjustment with allometry and intermediate outcomes. BMC Biol 15, 107 (2017). https://doi.org/10.1186/s12915-017-0448-5
Published:
DOI: https://doi.org/10.1186/s12915-017-0448-5