Abstract
A transfer learning system was designed to predict Xylosma racemosum compression strength. Near-infrared (NIR) spectral data for Acer mono and its compression strength values were used to resolve the weak generalization problem caused by using a X. racemosum dataset alone. Transfer component analysis and principal component analysis are domain adaption and feature extraction processes to enable the use of A. mono NIR spectral data to design the transfer learning system. A five-layer neural network relevant to the X. racemosum dataset, was fine-tuned using the A. mono dataset. There were 109 A. mono samples used as the source dataset and 79 X. racemosum samples as the target dataset. When the ratio of the training set to the test set was 1:9, the correlation coefficient was 0.88, and mean square error was 8.84. The results show that NIR spectral data of hardwood species are related. Predicting the mechanical strength of hardwood species using multi-species NIR spectral datasets will improve the generalization ability of the model and increase accuracy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Xylosma racemosum is widely distributed in Northeast China. Due to its density, high specific gravity, texture, and anti-corrosion and water resistance characteristics, the species is commonly used for furniture and structural material. Compression strength is one of the most important mechanical properties of tree species; however, traditional compression strength testing is time-consuming and costly (Rakotovololonalimanana et al. 2015). Furthermore, the species has natural heterogeneous or diverse polymer characteristics and mechanical parameters because of inner defects and other factors. Therefore, a single sample cannot represent the entire batch of boards accurately.
Near-infrared (NIR) spectroscopy is a nondestructive, economical and reliable approach to evaluate various properties of organic materials. The wavelength range of the NIR spectrum is 770–2500 nm and reflects the molecular hydrogen groups O–H, N–H, C–H vibrational information that illustrates their structure. Because NIR spectral absorption peaks differ for thevarious molecular hydrogen groups, complex materials and their physical and biological information can be chemically analyzed.
Various aspects of wood such as chemical components, mechanical properties, and degree of deterioration have been studied using NIR spectroscopy (Satoru and Hikaru 2015). The chemical absorption band reflects wood cellulose features that directly determine compression strength. Wood compression strength prediction models were successfully developed using NIR spectroscopy. Liang et al (2016) collected spectral data of 160 X. racemosum samples and designed a genetic algorithm backward interval partial least squares prediction model. When the ratio of calibration was 3:1, the model produced a 0.927 correlation coefficient. An artificial neural network (ANN) is also commonly used with NIR detection. Watanabe et al. (2014) compared the partial least squares regression model and the ANN prediction model with NIR spectroscopy and showed that ANN was more effective and accurate for wood NIR prediction.
However, in NIR spectroscopy analysis processing, spectral information is poor but may easily be covered by other information. On the other hand, the quantity and representativeness of the samples may be limited, thus limiting the ability of the prediction model to generalize and a limited scope of application. As a result, we proposed basing the transfer learning system on two species of hardwood data; NIR spectral data and corresponding compression strength values for A. mono were used to establish a compression strength prediction model for X. racemosum.
Transfer learning, an increasingly popular direction for machine learning research (Lu et al. 2015), transfers learned knowledge from a source domain to a target domain to establish a better model. Due to the small-scale, non-representative samples, useful features are obscured by large amounts of redundant information in the original data, which often leads to over-fitting and a narrow scope of application. With effective transfer learning algorithms and suitable source domain data, a system could produce more useful knowledge and thus better performance and generalization. Good results have been achieved in different areas such as computer vision, nature language recognition, and human behavior recognition. Wang and Mahadevan (2011) proposed a varied alignment approach for heterogeneous domain adaptation that utilizes natural image datasets to classify medical images. Yosinski et al. (2014) studied the transferability of features in deep neural networks utilized in language recognition and image recognition. Cook et al. (2013) reviewed the literature to highlight advances in transfer learning for human activity recognition.
In this study, we aimed to develop a transfer learning system to establish a X. racemosum compression strength prediction model. In wood microstructures, the effect of an S2 microfibril cell wall angle on wood compressive strength is significant. For example, microfibril angles of coniferous cell walls are much larger than those of normal wood, reaching about 45°, while the compressive strength of coniferous wood is only 50–60% that of normal wood (Li 2002). Because it is difficult for NIR spectroscopy absorbance to reflect the exact microfilament angle, A. mono was selected as a domain source because it microstructure is similar to that of X. racemosum. The spectral data of A. mono samples and corresponding compression strength values were considered a source dataset, and the spectral data of X. racemosum samples and corresponding compression strength values were the target dataset. In this transfer learning system, transfer component analysis (TCA) and principal component analysis (PCA) were regarded as an unsupervised learning representation to normalize the different spectral data for the two species. The source dataset was then used to pre-train a X. racemosum compression strength prediction model, and the target dataset was used to fine-tune this model. This prediction model should result in the acquisition of considerable information from the X. racemosum NIR spectral data, which have a high degree of accuracy, efficiency and a good generalization ability.
Materials and methods
Materials
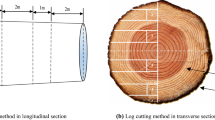
Six X. racemosum and six A. mono logs were collected from the Dailing Forestry Bureau, Heilongjiang Province. At 1.3 m on each log, 5-cm thick discs were cut. Following the Chinese National Standards, “Wood Physical and Mechanical Specimen Collection Methods (GB/T1936-2009)”, the discs were cut into standard 30 mm 20 mm 20 mm samples,76 for X. racemosum and 109 for A. mono. All samples were placed into a thermostat box at 22 °C with 12% moisture content and 65% relative humidity.
NIR spectral measurements
A NIR Quest (512) spectrometer (Ocean Optics, Weihai Optical Instrument Co., Ltd, Shanghai, China) provided 900–1700 nm spectra at 3-nm intervals to measure surfaces of the samples. Wavelengths from 1100 to 1700 nm provide information that is vital for the analysis of wood properties (Schimleck et al 2003; Todorović et al. 2015). A stable 512-pixel indium gallium arsenide array detector, located on a compact light pedestal with two-stage thermoelectric coolers and low-noise electronic components, scanned surfaces so that errors due to improper operation could be eliminated. After preheating 10 min and scanning the polytetrafluoroethylene reference tile for calibration, according to the “Standard Method for Near Infrared Qualitative Analysis of Wood (LY/T2053-2012)”, the detector scanned the surfaces of samples. The NIR spectrum for the samples was generated using SPEC view 7.1 (Insion Co., GmbH, Heilbronn, Germany) and exported to Excel (Microsoft, Redmond, WA, USA). Each sample was scanned 30 times and the data averaged. Figure 1 shows the process of NIR spectrum collection.
Mechanical parameters test
In accordance with Chinese National Standards, “Method of Testing Physical and Mechanical Properties of Wood (GB/T 15,780–1995)”, compression strength was measured using an electronic universal mechanical testing machine, loading at a constant rate. At a particular load, the sample was destroyed, the load was reduced, and the compression strength was recorded at that point.
Preprocessing of NIR spectra
For reducing any negative effects caused by incorrect operation during the collection of the raw NIR spectra, a standard normal variate (SNV) pretreatment and a Savitzky–Golay (SG) smoothing filter were used. SNV improves light scattering and Savitzky–Golay (SG) reduces high-frequency noise and spectrum baseline drift. SNV processing is:
where Xi is the original spectrum, µ is the mean of the original spectrum, and is the standard deviation of the original spectrum. The SG smoothing filter uses a polynomial approach to make a least square fit in moving windows. The total number of wavelength points per spectrum is D, the wavelength point sequence number is j(j = 1,2…D), the width of moving windows 2 m + 1 (− m,− m + 1,…m − 1, m), and aj = {a0,a1,…ak} is the weight coefficient that conforms to the k-order polynomial.
where is the NIR wavelength absorbance point of the moving window. In the moving window, the minimized error between the NIR spectra fitted by polynomials and original NIR spectra is:
Set \(\frac{\partial \varepsilon }{{\partial a_{j} }} = 0\), calculating the corresponding weight coefficient combination, when the smallest error occurs in different size windows.
Transfer learning system for Xylosma racemosum compression strength prediction
In the machine learning model, the limited input data does not adequately represent the species, and this type of model usually has poor generalization quality. With a suitable source dataset and suitable transfer learning approaches, a source domain’s knowledge and features could be transferred to a target task and help improve the model’s performance and generalization ability.
In this study, a transfer learning system was designed to establish a X. racemosum compression strength prediction model coupled with an A. mono sample dataset (Fig. 2).
The transfer component analysis (TCA) is a good domain adaptation algorithm and suitable for handling large datasets and prediction models (Pan et al. 2010). A. mono NIR spectral data was set as source domain and X. racemosum NIR spectral data as target domain. The source domain NIR spectral data (Ds) is denoted as:
where \(x_{{S_{1} }} \in \chi\) is the input and \(y_{{S_{1} }} \in \gamma\) isthe corresponding output. Similarly, the target domain data (DT) is denoted as:
P(XS), Q(XT) are the marginal distribution of XS and XT, respectively, and where P(XS) ≠ Q(XT), source data cannot be used to help target domain research. The TCA assists in finding a nonlinear map that gives good representation in a subspace between source and target domain (and minimizes their marginal distribution). With this method, P(XS) P(XT). Because of the strong correlation between the two domains, it is assumed that P(YSXS)P(YTXT). (Source data could be used to help target task).
The TCA uses maximum mean discrepancy (MMD) (Borgwardt et al. 2006) to define the marginal distribution after mapping to minimize the distance shown in Eq. 6, where \(\varphi\) is a nonlinear map, to map the source data and target data in a reproducing Kernel Hilbert space.
Because it is difficult to solve for \(\varphi\), the TCA introduces a kernel trick to transfer Eq. 6 as:
where (n1 + n2) × (n1 + n2) is a kernel matrix, \(K_{S,S} ,\;K_{T,T} ,\;K_{T,S}\) respectively, are the kernel matrices defined by k on the data in the source domain (src), target domain (tar), and cross domains, and L = [Lij] ≥ 0 with:
Kernel \(k(\cdot , \cdot)\) may be solved through learning the kernel matrix \(k\). The TCA uses a novel kernel learning method to transfer kernel K to a low-rank presentation m-dimension W (Pan et al. 2010) shown in Eq. 10:
The solution of W is the final solution of the TCA for redundant information in the NIR spectral data that will have a negative effect on the model’s performance and generalization ability, in this study, we also used a principal component analysis (PCA) to eliminate redundant information. The PCA could find the eigenvectors corresponding to the largest eigenvalues of the covariance matrix of the data to reduce the dimensions of the data. These values could be from n to d, where d < n. According to the d-dimension information-proportion of the original data, d can be solved (Wold et al. 1987).
The dataset for Acer mono after feature selection was input into a 5-layer learning neural network for training, which stopped when the value of the loss function kept oscillating within a certain step length.
Xylosma racemosum samples were randomly divided as a training set and a test set. The training set was input into the model for fine-tuning. The test set was used to evaluate the prediction model’s performance. Because the parameters in the model had been learned before, the learning rate was set as one-fifth of the original. We used 10 trainings and obtained the average model performance.
Model evaluation standard
By comparing the NIR wood property prediction model, we evaluated the transferability of wood NIR spectroscopy knowledge, considering not only the accuracy and data dependence of the model but also the generalization ability of the model. Statistical measurements (Schimleck et al. 2003), mean square error (MSE) and determination coefficient (r) are commonly used in NIR model evaluation. A good model has higher determination coefficients and lower mean square errors. Furthermore, the performance of the model is evaluated under different training and prediction sets, which reflect the generalization ability of the model.
Results and discussion
Determination of compression strength
Compression strength values of 109 A. mono samples and 76 X. racemosum samples obtained by the mechanical testing machine are shown in Table 1. The detailed distribution of the compression strength values are shown in Fig. 3.
Near-infrared spectroscopy and spectra pre-processing
The raw NIR spectra of samples were collected from 899.77 to 1720.81 nm, and 512 NIR spectral wavelength points were obtained (Fig. 4). The raw NIR spectral data were then pre-processed using SNV and SG smoothing filters (Figs. 5 and 6).
Compared with the spectra in Fig. 4, in Figs. 5 and 6, light scattering was lessened and the trend of change was more uniform after SNV processing. The main absorption peaks were also more obvious and the spectral profiles more regular after SG smoothing.
Transfer learning prediction model performance evaluation and analysis
The A. mono dataset was used as a source domain and the X. racemosum dataset as the target domain. Input for both source domain and target domain the 512 dimensional NIR spectral data for TCA domain adaptation processing. Two 50-dimensional feature matrices were produced. Inputting 50-dimensional feature matrix for the two kinds of timber for PCA, set the loss rate less than 5%, then 20-dimensional features matrix of two kinds of timber NIR data were obtained.
A five-layer neural network was established in Keras, an open-source neural network library. All spectral data and corresponding compression strength values of the 109 A. mono samples were inputted to pre-train the prediction model. The X. racemosum data and corresponding compression strength values were divided into an eight-group training set and a test set. Training sets were chosen randomly to account for 10%, 20%,…, 80% of the total X. racemosum samples, and the rest were used for the test set. This eight-group dataset was input into the prediction model, respectively, whose parameters had been previously learned by the source dataset. After 100 epochs, the loss function of the model was generally stable. The model’s predictive performance is shown in Table 2.
The model yielded 8.36 MSE and 0.88 r for predictive performance even if the training sets only accounted for 10% in total samples of X. racemosum used for the compression strength. Under the ratio of 60% training set with 40% test set, 5.23 MSE and 0.94 r predictive performance were obtained by the prediction model.
For testing the effect of different parts in this transfer learning system, three controlled trials were designed by cutting some wood sections. For the first trial, the TCA and pre-training process were cut. With the second trial, only the TCA section was cut, and for the third, only the pre-training section was cut.
The effects of the different processing methods and models, based on the model evaluation criteria, are shown in Figs. 7 and 8. The blue line represents a complete transfer learning system, the light blue line represents a system lacking TCA, the black line a system lacking the pre-training process, and the red line a system lacking both the TCA and pre-training process.
As shown in Figs. 7 and 8, with TCA processing, the prediction model had a better performance and generalization ability because common features could be found that were implicitly expressed in the mapped spectral matrix. After effective extraction, common hardwood features reflected by NIR spectra were refined, and much redundant information was eliminated. This step was very useful in the following supervised learning task.
Conversely, without domain adaption, when the source dataset was directly input into the prediction model for pre-training, there was poor prediction because the model is insensitive to input target data. Because the NIR spectral data for the two specimens did not share some marginal distribution, only certain learned knowledge was useful. Along with the increase in target data, the knowledge learned from the source data has a negative transfer effect.
The complete transfer learning prediction model has better performance and transformation ability (Figs. 7 and 8). In addition, after pre-training, the model only required, on average, about 100iterations to accomplish the training process, so it was 10 times faster than the prediction model without the transfer learning processing.
The use of the pre-trained model and transfer learning may be considered a conceptual drift process, which could conduct automatic regularization. This process guided the neural network function to converge into a specific area containing the hardwood knowledge and to exclude noise caused by the limitation of input data. This automatic regularization process made the gradient descent direction of the loss function more explicit, reduced the oscillation of the descent direction, accelerated the convergence speed of the loss function, and reduced the training time of the model. Therefore, this transfer learning model has strong generalization ability, good performance and is efficient.
Conclusions
A transfer learning system based on NIR spectral data was developed. With A. mono data, the compression prediction model established for X. racemosumhas good performance and strong generalization ability.
Both A. mono and X. racemosum are hardwoods. There is a general knowledge of mechanical strength of hardwood in their NIRspectral data, which can be extracted by the domain adaptation method.
The spectral characteristic matrix contains the interrelation between wood characteristics and compression strength, which is implicitly expressed in the hyperparameters of the model. This a priori knowledge may be transferred to the target model and thus improve its performance, generalization ability and efficiency.
By means of transfer learning and multi-species spectral data, the mechanical strength of a hardwood can be accurately predicted.
References
Borgwardt KM, Gretton A, Rasch MJ, Kriegel HP, Schölkopf B, Smola AJ (2006) Integrating structured biological data by kernel maximum mean discrepancy. Bioinformatics 22(14):e49–e57
Cook D, Feuz KD, Krishnan NC (2013) Transfer learning for activity recognition: a survey. Knowl Inf Syst 36(3):537–556
Li J (2002) Wood Science, 2nd edn. Higher Education Press, Beijing, pp 304–341
Liang H, Cao J, Tu W, Lin X, Zhang Y (2016) Nondestructive determination of the compressive strength of wood using near-infrared spectroscopy. BioResources 11:7205–7213
Lu J, Behbood V, Hao P, Zuo H, Xue S, Zhang G (2015) Transfer learning using computational intelligence: a survey. Knowl-Based Syst 80:14–23
Pan SJ, Tsang IW, Kwok JT, Yang Q (2010) Domain adaptation via transfer component analysis. IEEE Trans Neural Netw 22(2):199–210
Rakotovololonalimanana H, Chaix G, Brancheriau L, Ramamonjisoa L, Ramananantoandro T, Thevenon MF (2015) A novel method to correct for wood MOE ultrasonics and NIRS measurements on increment cores in Liquidambar styraciflua L. Ann For Sci 72(6):753–761
Satoru T, Hikaru K (2015) A review of recent application of near infrared spectroscopy to wood science and technology. J Wood Sci 61:213–220
Schimleck LR, Mora C, Daniels RF (2003) Estimation of the physical wood properties of green Pinus taeda radial samples by near infrared spectroscopy. Can J For Res 33(12):2297–2305
Todorović N, Popović Z, Milić G (2015) Estimation of quality of thermally modified beech wood with red heartwood by FT-NIR spectroscopy. J Wood Sci Technol 49(3):527–549
Wang C, Mahadevan S (2011) Heterogeneous domain adaptation using manifold alignment// IJCAI 2011. In: Proceedings of the 22nd international joint conference on artificial intelligence, Barcelona, Catalonia, Spain, July 16–22, 2011. DBLP, 2011
Watanabe K, Kobayashi I, Matsushita Y, Saito S, Kuroda N, Noshiro S (2014) Application of near-infrared spectroscopy for evaluation of drying stress on lumber surface: a comparison of artificial neural networks and partial least squares regression. Dry Technol 32(5):590–596
Wold S, Esbensen K, Geladi P (1987) Principal component analysis. J Chemom Intell Lab Syst 2(1–3):37–52
Yosinski J, Clune J, Bengio Y, Lipson H (2014) How transferable are features in deep neural networks? In: Advances in neural information processing systems 27 (NIPS’ 14), NIPS Foundation, 2014, 3320–3328. https://papers.nips.cc/paper/5347-how-transferable-are-features-in-deep-n%E2%80%A6.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Project funding: The project is fully funded by the Program of National Natural Science Foundation of China (CN) (31700643) and Fundamental Research Funds for the Central Universities (2572015AB24).
The online version is available at http://www.springerlink.com.
Corresponding editor: Yu Lei.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Shi, G., Cao, J., Li, C. et al. Compression strength prediction of Xylosma racemosum using a transfer learning system based on near-infrared spectral data. J. For. Res. 31, 1061–1069 (2020). https://doi.org/10.1007/s11676-019-01052-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-019-01052-2