Abstract
Adipogenesis is important in the development of fat deposition. Evidence showed that plant microRNAs (miRNAs) could be absorbed by the digestive tract and exert regulatory effects on animals’ physiological processes. However, the regulation of plant miRNA on host lipogenesis remains unknown. This study explored the potential function of plant miRNA, miR167e-5p, in adipogenesis in vitro. The presentation of plant miR167e-5p improved lipid accumulation in 3T3-L1 cells. Bioinformatics prediction and luciferase reporter assay indicated that miR167e-5p targeted β-catenin. MiR167e-5p could not only negatively affect the expression of β-catenin but also showed a positive effect on several fat synthesis–related genes, peroxisome proliferator–activated receptor gamma (Pparγ), CCAAT/enhancer-binding protein α (Cebpα), fatty acid-binding protein 4 (Ap2), lipolysis genes, adipose triglyceride lipase (Atgl), and hormone-sensitive lipase (Hsl) messenger RNA levels. Meanwhile, lipid accumulation and the expression of the β-catenin and other five fat synthesis–related genes were recovered to their original pattern by adding the miR167e-5p inhibitor in 3T3-L1 cells. The immunoblot confirmed the same expression pattern in protein levels in β-catenin, PPAR-γ, FAS, and HSL. This research demonstrates that plant miR167e-5p can potentially affect adipogenesis through the regulation of β-catenin, suggesting that plant miRNAs could be a new class of bioactive ingredients in adipogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Adipose tissue is the basis of energy metabolism in animals. Its function as an essential endocrine organ is closely related to metabolic diseases such as diabetes (Perry et al. 2017), cardiometabolic disease (Scheja and Heeren 2019), inflammation (Yamashita et al. 2018), and microvascular pathology (Yamashita et al. 2018). Adipocytes are the cells that primarily compose adipose tissue, the primary energy storage as fat (Fruhbeck et al. 2001). Normal metabolism of fat body is required for the proliferation and differentiation of adipocytes to satisfy an organism’s energy needs (Kahn et al. 2006; Yang and Mottillo 2020). In the animal husbandry field, it is reported that adipose tissue can improve the quality of meat, such as the taste, juiciness, and softness of the meat, by enhancing intramuscular fat concentration (Pietruszka et al. 2015). Studies showed that many genes were involved in adipogenesis, including peroxisome proliferator–activated receptor-γ (PPARγ), CCAAT enhancer-binding protein-α (C/EBPα), adipose triglyceride lipase (Atgl), and hormone-sensitive lipase (Hsl), among others (Li et al. 2022). These genes affect adipogenesis through the mutual transmission network of information, like the Wnt signaling pathway (Jun et al. 2020; Martinez Calejman et al. 2020; Wang et al. 2020; Guo et al. 2022). These regulations of adipogenesis are initiated by the sequential repression of particular genes by some critical regulatory molecules, such as microRNAs ( Shen et al. 2016; Hao et al. 2019; Qi et al. 2021).
MicroRNA (miRNA) is a small single-stranded non-coding RNA molecule (~ 21 nucleotides) identified in plants, animals, and some viruses that functions in RNA silencing and post-transcriptional regulation of gene expression (Ha and Kim 2014). It has been revealed that it is widely implicated in adipogenesis by inhibiting particular genes in the mammalians (Shen et al. 2016; Hao et al. 2019; Qi et al. 2021). Current studies have shown that exogenous miRNAs can regulate gene expression in cross-kingdom. Host uptake of dietary and orally administered miRNAs can be absorbed into the body through the SIDT-1 gene of the stomach (Zhou et al. 2020b; Chen et al. 2021a). They may also cross the intestinal barrier and enter the blood (Ju et al. 2013; Mu et al. 2014; Luo et al. 2017). These miRNAs subsequently enter the tissue to involve in physiological and pathological processes by regulating gene expressions in hosts, such as suppressing influenza viruses (Zhou et al. 2015), replicating novel coronavirus (Zhou et al. 2020a), tumor growth (Chin et al. 2016), caste development (Zhu et al. 2017), atherosclerosis (Hou et al. 2018), and intestinal development (Li et al. 2019a, 2019b). The potential of miRNAs’ cross-kingdom transferring and gene regulation should be considered one of the plants’ valuable characteristics in human health and animal husbandry. Chinese herbs have been found to contain miRNA, which has been proven to have therapeutic impacts on illness (Zhou et al. 2020a, 2020b). Some miRNAs in Gmelina arborea (Dubey et al. 2013) and Curcuma longa (Lukasik and Zielenkiewicz 2014) were discovered to hybridize with the various targets of signal transduction and apoptosis that play a potential role in preventing diseases such as diabetes mellitus type 2, blood-borne disease, and other urinary infections (Lukasik and Zielenkiewicz 2014). Oral administration of exosome-like nanoparticles from grapes containing abundant miRNAs aids in protecting mice from dextran sulfate sodium (DSS)-induced colitis via induction of intestinal stem cells through the Wnt/β-catenin pathway (Ju et al. 2013). Plant miRNAs derived from various sources, like miR167e-5p, have been found in animals’ adipose tissue (Luo et al. 2017) and were previously identified as a regulator of the Wnt signaling pathway (Li et al. 2019a), which means that they may represent a potential approach to regulate adipogenesis. Further data and research are necessary to completely comprehend its possible influence on adipogenesis. Plant miRNA may help the knowledge of the molecular processes behind Chinese herbal medicine and give preventative methods and safer, more “natural” therapies for metabolic diseases or improve the level of livestock production.
In this study, we explored the potential involvement of a plant miR167e-5p in fat metabolism through the β-catenin pathway with a mouse 3T3-L1 pre-adipocyte cell line, which has been widely used as an adipocyte differentiation model in animal studies. The results from this study shed light on the interaction of exogenous plant miRNA with adipogenesis and its implicit as a new class of components for regulating adipogenesis in the future.
Materials and methods
Cell culture and transfection of cells with miR167e-5p
Murine pre-adipocyte 3T3-L1 cells were procured from the American Type Culture Collection (ATCC Manassas, VA) agents (Beijing Zhongyuan Co., Ltd., Beijing, China). The 3T3-L1 cells were maintained in DMEM with high glucose (1% antibiotic/antimycotic solution (Gibco, Grand Island, NY) and 10% fetal bovine serum (Gibco)) at 5% CO2 and 37 °C. The scrambled negative control oligonucleotides, mature plant miR167e-5p mimics (mature sequence: UGAAGCUGCCAGCAUGAUCUG), and miR167e-6p antisense chain were synthesized by RiboBio (Guangzhou, China). A 12-well plate was seeded with 3T3-L1 cells at 5 × 104 cells/well overnight (70–80% confluency), then transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instruction. The treatment groups were as follows: negative control (NC) group (20 pmol/well, scrambled negative control oligonucleotides), miR167e-5p group (20 pmol/well, plant miR167e-5p mimics), and miR167e-5p + inhibitor group (20 pmol/well miR167e-5p mimics + 40 well pmol miR167e-5p antisense chain) (n = 6). To induce cell differentiation, cells in different groups were incubated with DMEM high-glucose medium supplemented with inducers at 0.5 mM isobutyl methylxanthine (IBMX; Sigma, Darmstadt, Germany), 0.5 mM dexamethasone (Sigma), and 20 nM insulin (Sigma) for 2 d, followed by further differentiation with DMEM high-glucose medium plus 20 nM insulin, then changed the medium with 20 nM insulin every 2 d until day 8. There were six biological repeats per group.
Oil Red O staining
The differentiated cells were washed with PBS and then fixed with 4% paraformaldehyde for 30 min at room temperature (RT). The fixed cells were incubated with 0.5 mL of 1% filtered Oil Red O for 30 min at RT. Cells were washed two times with PBS, and the strained fat droplets in the cells were visualized and photographed under light microscopy (Nikon TE2000, Tokyo, Japan). Further, isopropanol (200 μL/well) was used to extract cellular Oil Red O and detect optical absorbance at 490 nm for quantification analysis. There were three biological repeats per group.
Triglyceride assay
The differentiated cells were rinsed with PBS, and 100 μL of lysis buffer was added to each well. The cell lysates’ triglyceride contents were assayed by Food Triglyceride Assay Kit (Applygen, Beijing, China); total protein per sample was measured by a standard BCA method (Protein Detection Kit; BioTeke Corporation, Beijing, China) for normalization of triglyceride concentration. According to the protocol provided in the kit, the assays were performed in 96-well plates, and readouts were obtained on a microplate reader (Thermo Labsystems MK3; Thermo Fisher Scientific, Inc., Waltham, MA) at a wavelength of 510 nm. There were five biological repeats per group.
RNA isolation
The total RNA of cell samples was extracted using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. The quantity and quality of RNA were evaluated on an ND-2000 NanoDrop Spectrophotometer (NanoDrop Technology, Wilmington, DE) and 0.8% agarose gel electrophoresis. All prepared RNA samples were stored in a − 80 °C freezer for downstream analysis. There were six biological repeats per group.
QPCR analyses
Total RNA (2 μg) was reverse-transcribed for complementary DNA (cDNA) synthesis by using M-MLV Reverse Transcriptase kit (Promega, Madison, WI) with OligodT18 (for messenger RNA (mRNA)) or a specific stem-loop RT primer (for miR167e-5p). The miRNA- and mRNA-specific primers were designed with Premier Primer 5.0 (Premier Software, San Francisco, CA) and synthesized at Sangon Biotech (Shanghai, China) (Table 1). qPCR was performed on a Bio-Rad CFX Manager 3.1 instrument (Applied Biosystems, Waltham, MA) with U6 as an internal control for miRNA or Gapdh for mRNA. Reactions contained 2 μL cDNA (fivefold diluted), 10 μL 2 × GoTaq qPCR Master Mix (Promega), and 0.4 μL of each primer (10 Μm). The thermal profile of real-time PCR involved an initial denaturation step at 95 °C for 5 min, followed by 40 cycles at 94 °C for 15 s, 15 s at the corresponding annealing temperature extension at 72 °C for 30 s, followed by a quick denaturation at 95 °C for 10 s, plus a slow ramp from 60 to 95 °C to generate a melting curve to confirm the specificity of the amplified product. A negative control without a template was included in qPCR for both miRNA and mRNA. All reactions were performed in triplicate. The 2−ΔΔCt method was employed for the expression analysis. There were six biological repeats per group.
Western blot analysis
Total protein was isolated from 3T3-L1 cells using RIPA buffer with 1 nM phenylmethylsulfonyl fluoride (PMSF) and quantified with a BCA Total Protein Assay Kit (Thermo Fisher). The 30 μg protein per sample was supplemented with β-mercaptoethanol boiled for 10 min, resolved on 5% and 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), respectively, then transferred to a polyvinylidene difluoride (PVDF) membrane. The PVDF membranes were probed with primary antibodies against human β-catenin (1:500, D260137; Sangon Biotech), PPAR-γ (1:1000; CST, Danvers, MA), FAS (1:1000; CST), and ATGL (1:1000; CST), followed by HRP-conjugated secondary antibody. GAPDH (1:50,000; Bioworld Technology, St. Louis Park, MN) was served as a control for equal loading. The probed PVDF membranes were evaluated using a multicolor fluorescence chemiluminescence gel imager (ProteinSimple, San Jose, CA). The densitometry of the band was measured using ImageJ and normalized to GAPDH. There were six biological repeats per group.
Bioinformatics analysis
The NCBI GenBank database was used to get full-length cDNAs of the pig/human/mouse genes. MiR167e-5p-matched locations in the full CDS/UTR of the transcripts were analyzed by miRanda and TargetScan software. The software utilized some common parameters to identify whether a transcript was a miR167e-5p target. The initial “seed rules” criteria for target identification were base pairing between the “seed” (the core sequence including the first 2 to 8 bases of the mature miRNA) and the target (Smalheiser and Torvik 2006).
Dual-Luciferase Reporter Assay
Based on the predicted miRNA-mRNA binding sequences, normal sequences bearing a miR167e-5p seed binding site or with the binding site of miR167e-5p on the β-catenin 3′-UTR deleted were generated by two complementary chemically synthesized primers (Sangon Biotech) as follows: Wt-mmu-β-catenin-3′-UTR-sense TCGAGTTGTATCTAAAGTCCGGTGTTGCCAGCTTCAGTTGGTTCCTGT, Wt-mmu-β-catenin-3′-UTR-antisense CTAGACAGGAACCAACTGAAGCTGGCAACACCGGACTTTAGATACAAC Del-mmu-β-catenin-3′-UTR-sense TCGAGTTGTATCTAAAGTCCGGTGTTGCAGTTGGTTCCTGT, and Del-mmu-β-catenin-3′-UTR-antisense CTAGACAGGAACCAACTGCAACACCGGACTTTAGATACAAC. The typical 3′-UTR sequence with the miR167e-5p binding site (wild type, Wt) and the 3′-UTR sequence with the deleted miR167e-5p binding site (deleted, Del) of β-catenin gene were cloned into luciferase reporter plasmids, respectively. In brief, the complementary oligonucleotides were resuspended with a ratio at 1:1 (1 mg/μL each) in annealing buffer (10 mM Tris, pH 7.5 ~ 8.0, 50 mM NaCl, 1 mM EDTA) and heated at 95 °C for 10 min. The temperature was then gradually lowered to room temperature. Annealed products were cloned into the pmirGLO vector (Promega) downstream from the firefly luciferase coding region (between XhoI and XbaI sites). HeLa cells were seeded in 96-well cell culture plates (4 × 104 cells per well) and cultured in RPMI 1640 (Life Technologies, Grand Island, NY) with 10% fetal bovine serum (FBS). On the next day, the recombinant pmirG′O-3′-UTR vector (100 ng/well) mixed with their corresponding miR167e-5p mimics or NC (3 pmol/well; RiboBio) was transfected to the seeded cells with Lipofectamine 2000 (Invitrogen) by following a recommended procedure. Cells were harvested at 24 h after transfection, and the luciferase activity was detected using a Dual-Luciferase Reporter Assay kit (Promega) according to the recommended manufacturer’s instruction. The firefly luciferase activity (firefly luciferase activity/Renilla luciferase activity) was normalized and compared with the pmirGLO vector for its relative activity. There were eight biological repeats per group.
Statistical analysis
The ANOVA with the least significant difference (LSD) test was used for multiple comparisons (the same superscripts denoted a not-significant difference (P > 0.05), and different superscripts denoted a significant difference (P < 0.05); *P < 0.05 was considered statistically significant). Data were presented as mean ± standard error of the mean (SEM), and all data analyses were performed via GraphPad Prism software 8.0 (GraphPad Software, San Diego, CA).
Results
Plant miR167e-5p regulates adipogenesis in 3T3-L1 cells.
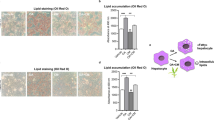
To investigate the role of miR167e-5p in adipogenesis, 3T3-L1 cells were transfected with synthetic miR167e-5p mimics and treated with differentiation inducers to induce differentiation. The development of lipid droplets was observed using Oil Red O after 8 d. The miR167e-5p mimic group demonstrated a significant increase in lipid droplets in 3T3-L1 cells (P < 0.05), as compared to that in the control (NC) and the group with miR167e-5p + inhibitor (Fig. 1A, B; P < 0.05). A similar pattern of change was observed in the triglyceride assay; i.e., the triglyceride levels in cells were considerably higher in the miR167e-5p mimic group, whereas they were lower in the NC group and miR167e-5p + inhibitor group (Fig. 1C, P < 0.05).
Plant miR167e-5p regulates adipogenesis in the 3T3-L1 cells. 3T3-L1 cells were transfected with negative control (NC), miR167e-5p, and miR167e-5p + miR167e-5p inhibitor groups and treated with differentiation inducers for 8 d. (A, B) Quantification of lipid droplets by staining with Oil Red O (n = 3, scale bars, 200 px). (C) Triglyceride concentration in adipocytes and adjusted by protein content (n = 5). The same superscripts denoted a not-significant difference (P > 0.05); different superscripts denoted a significant difference (P < 0.05).
Plant miR167e-5p directly targets β-catenin.
With miRanda and TargetScan software, the analyzed results showed that miR167e-5p potentially targets β-catenin, a key molecule in the Wnt/β-catenin pathway. A putative conserved binding site was detected in β-catenin from various species (Fig. 2A). We calculated the minimum free energy of binding to be less than − 20 kcal/mol. Luciferase reporter assay showed that the luciferase activity of the Wt group was significantly reduced to nearly 50%, compared to luciferase activity in the Del group after being exposed to miR167e-5p mimics (Fig. 2B, P < 0.05).
MiR167e-5p targets β-catenin. (A) The predicted target β-catenin 3′-UTR and miR167e-5p among different species. Construction of the luciferase reporter plasmid carrying the wild-type (Wt) or Delete (Del) miR167e-5p complementary site. (B) Luciferase activities in HeLa cells co-transfected with luciferase reporters described in (A) and miR167e-5p mimics or NC (firefly luciferase activity/Renilla luciferase activity, *P < 0.05, n = 8).
Evaluation of gene expression in 3T3-L1 cells transfected with plant miR167e-5p.
To pinpoint changes in adipogenesis affected by miR167e-5p, we evaluated the changes of expression of 5 fat marker genes, plus β-catenin. As expected, the expression of the target gene β-catenin was affected negatively (Fig. 3B, P < 0.05) after exposure to miR167e-5p (Fig. 3A, P < 0.05). The reduction was recovered by adding a miR167e-5p inhibitor (Fig. 3B, P < 0.05), which indicated that miR167e-5p probably could affect adipogenesis in cells through the degradation of β-catenin mRNA. As shown in Fig. 3C–E, the relative expression was upregulated in the fat marker genes Cebpα, Pparγ, and Ap2 compared to the control group (P < 0.05). In our reverse experiment, the expression of these three genes was recovered after the addition of inhibitor in the miR167e-5p group (P < 0.05). The same significant upregulation pattern was observed in two lipolysis-related genes, Atgl and Hsl, in the miR167e-5p group (P < 0.05), even though the expression failed to restore the inhibitor group after the addition of the inhibitor (Fig. 3F, G).
MiR167e-5p suppresses β-catenin and promotes fat-related gene expression. The 3T3-L1 cells were transfected with NC, miR167e-5p, and miR167e-5p + miR167e-5p inhibitor and treated with differentiation inducers for 8 d. (A) qPCR analysis of relative miR167e-5p levels. (B–G) qPCR analysis of relative fat-related gene mRNA levels. The same superscripts denoted a not-significant difference (P > 0.05); different superscripts denoted a significant difference (P < 0.05). ND: no detected, n = 6.
Protein expression in 3T3-L1 cells after transfected with plant miR167e-5p.
To further confirm the function of miR167e-5p on adipogenesis, we detected the protein expression levels of β-catenin, PPAR-γ, FAS, and ATGL by Western blot. The result showed that the expression of β-catenin was reduced in the miR167e-5p-over-expressed group, and the inhibitor could restore this reduction (Fig. 4A, B; P < 0.05). On the contrary, PPAR-γ, FAS, and ATGL protein expression levels were increased by miR167e-5p over-expression, and the inhibitor diminished this increment (Fig. 4A, C–E; P < 0.05).
Plant miR167e-5p suppressed β-catenin and increased fat-related protein expression. Western blot was used to evaluate β-catenin (A, B), PPAR-γ (A, C), FAS (A, D), and ATGL (A, E) protein levels after transfection with nucleic acids (NC)/miR167e-5p/miR167e-5p + inhibitor (n = 6; LSD tests). The same superscript denotes a not-significant difference (P > 0.05); a different superscript denotes a significant difference (P < 0.05).
Discussion
Adipocyte has a much more unexpected “native collar.” It releases a wide range of protein factors and signals termed adipokines, fatty acids, and other lipid moieties linked to energy balance and the inflammatory response (Trayhurn 2005). Genetic evidence in humans connects several Wnt pathway members to body fat distribution, obesity, and metabolic dysfunction, suggesting that this pathway also has functions in adipocytes (Bagchi et al. 2020). Canonical Wnt/β-catenin signaling is a well-studied endogenous regulator that inhibits adipogenesis and reduces adipocyte deposits. Activation of the Wnt/β-catenin signaling pathway inhibits adipogenesis, while disruption of Wnt signaling leads to spontaneous adipogenesis. Stabilizing free cytosolic β-catenin can activate the Wnt/β-catenin signaling pathway and further repress adipogenesis by preventing the induction of downstream genes, PPAR-γ and C/EBP-a, along the pathway (Jeon et al. 2016). In 3T3-L1 adipocytes, this lipid accumulation and inhibited fat synthesis factors accompanied the induction of β-catenin, effectively reversed by the β-catenin inhibitor (Shen et al. 2019). Our study is consistent with previous results that suppression of β-catenin expression increased the expression levels of PPAR-γ, C/EBP-α, and FAS in 3T3-L1 cells. Furthermore, our results also show that the increased fat synthesis is accompanied by increased fat decomposition, in which ATGL and HSL are responsible for TG’s breakdown in adipose tissue, and miR167e-5p significantly up-regulated these critical enzymes. Inhibition of plant miR167e-5p by small molecules substantially restored the fat synthesis genes, Cebpα, Pparγ, and Ap2, even though the mRNA expression of Atgl and Hsl was restored unsuccessfully by the inhibitor. The inhibitor can only competitively bind to miR167e-5p partially unless a dosage of inhibitor beyond cell capacity is provided. Protein results confirmed that miR167e-5p promoted fat metabolism. These findings re-emphasized the data above that miR167e-5p probably enhances fat synthesis accompanied by an increase in adipolysis by inhibiting the β-catenin expression. That assures that fat synthesis and adipolysis are always in a dynamic balance state to co-regulate adipogenesis in the body under physiological conditions (Pistor et al. 2015). In addition, adipogenesis is a complicated physiological process that involves several genes, intimate linkage and interconnection, and the inter-regulated message network (Jun et al. 2020; Martinez Calejman et al. 2020; Wang et al. 2020; Guo et al. 2022).
The mechanism by which Chinese herbal medicine treats diseases has remained a mystery. The research on cross-kingdom regulation of plant miRNAs and the study of influenza and novel coronavirus inhibition by honeysuckle miR2911 provide new insights into the disease-treating mechanisms of herbal medicine (Zhou et al. 2020a, 2020b). We have discovered that plant miR167e-5p can directly target β-catenin to promote adipogenesis, which plays a critical role in the Wnt/β-catenin signal pathway. MiR167e-5p downregulates the β-catenin expression to inhibit Wnt/β-catenin signaling and promotes 3T3-L1 preadipocyte adipogenesis. This finding is also in agreement with the reports that the β-catenin gene activates the canonical Wnt/β-catenin pathway to inhibit the crucial adipogenic transcription factor PPAR-γ and C/EBP-α expression to suppress adipogenesis (Yamashita et al. 2018; Guo et al. 2022). The results bring broad prospects to explore more plant-derived miRNAs that could affect fat regulation, exploring their mechanisms to keep fat content at a reasonable level, considering health features for humans. However, the concept of plant miRNA regulation in cross-kingdom is relatively new in adipogenesis and health, and we are just beginning to define the individual steps in this process.
Many Chinese herbs and woody plants have been frequently used in animal husbandry to increase output. Mulberry leaf powder in finishing pigs’ diets increased growth performance, as evidenced by higher final body weight, and improved carcass attributes, as evidenced by higher slaughter weight, carcass weight, carcass yield, and fatty acid composition in pork (Chen et al. 2021b). Diet-derived plant miRNA has been found in abundance in numerous tissues, including fat and muscle (Luo et al. 2017), which might be essential components regulating meat quality. Research on plant miRNAs linked to meat development can help livestock become leaner, store less fat, and improve the carcass’s quality and the meat’s flavor. A previous study has found that 16 maize miRNAs, including miR167e-5p in pig abdominal fat and muscle, exhibited relatively high abundance (Luo et al. 2017). Our data demonstrated that plant miR167e-5p overexpression increased droplet formation and triglyceride content to promote lipid accumulation. The phenotype mentioned above was significantly decreased when miR167e-5p was inhibited by a plant miR167e-5p inhibitor. All those results imply that plant miR167e-5p can induce adipogenesis. It is favorable for developing and using woody feed in livestock production or Chinese herbal herbs in illness treatment.
Conclusions
In conclusion, we discovered that miR167e-5p showed a positive effect on adipogenesis that promotes fat metabolism by targeting β-catenin and further blocking metabolite pathway. Our findings shed light on the role of plant miRNA in adipocyte biology to be favorable for a potential to develop and use the woody feed in livestock production or Chinese herbs in illness treatment.
Data availability
The data that support the findings of this study are available from the corresponding author, M.L., upon reasonable request.
References
Bagchi DP, Nishii A, Li Z, DelProposto JB, Corsa CA, Mori H, Hardij J, Learman BS, Lumeng CN, MacDougald OA (2020) Wnt/beta-catenin signaling regulates adipose tissue lipogenesis and adipocyte-specific loss is rigorously defended by neighboring stromal-vascular cells. Mol Metab 42:101078
Chen Q, Zhang F, Dong L, Wu H, Xu J, Li H, Wang J, Zhou Z, Liu C, Wang Y, Liu Y, Lu L, Wang C, Liu M, Chen X, Wang C, Zhang C, Li D, Zen K, Wang F, Zhang Q, Zhang CY (2021a) SIDT1-dependent absorption in the stomach mediates host uptake of dietary and orally administered microRNAs. Cell Res 31(3):247–258
Chen Z, Xie Y, Luo J, Chen T, Xi Q, Zhang Y, Sun J (2021b) Dietary supplementation with Moringa oleifera and mulberry leaf affects pork quality from finishing pigs. J Anim Physiol Anim Nutr (berl) 105(1):72–79
Chin AR, Fong MY, Somlo G, Wu J, Swiderski P, Wu X, Wang SE (2016) Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res 26(2):217–228
Dubey A, Kalra SS, Trivedi N (2013) Computational prediction of miRNA in Gmelina arborea and their role in human metabolomics. Am J Biosci Bioeng 1:62–74
Fruhbeck G, Gomez-Ambrosi J, Muruzabal FJ, Burrell MA (2001) The adipocyte: a model for integration of endocrine and metabolic signaling in energy metabolism regulation. Am J Physiol Endocrinol Metab 280(6):E827-847
Guo J, Qian L, Ji J, Ji Z, Jiang Y, Wu Y, Yang Z, Ma G, Yao Y (2022) Serpina3c regulates adipose differentiation via the Wnt/beta-catenin-PPARgamma pathway. Cell Signal 93:110299
Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15(8):509–524
Hao W, Liu HZ, Zhou LG, Sun YJ, Su H, Ni JQ, He T, Shi P, Wang X (2019) MiR-122-3p regulates the osteogenic differentiation of mouse adipose-derived stem cells via Wnt/beta catenin signaling pathway. Eur Rev Med Pharmacol Sci 23(9):3892–3898
Hou D, He F, Ma L, Cao M, Zhou Z, Wei Z, Xue Y, Sang X, Chong H, Tian C, Zheng S, Li J, Zen K, Chen X, Hong Z, Zhang CY, Jiang X (2018) The potential atheroprotective role of plant MIR156a as a repressor of monocyte recruitment on inflamed human endothelial cells. J Nutr Biochem 57:197–205
Jeon M, Rahman N, Kim YS (2016) Wnt/beta-catenin signaling plays a distinct role in methyl gallate-mediated inhibition of adipogenesis. Biochem Biophys Res Commun 479(1):22–27
Ju S, Mu J, Dokland T, Zhuang X, Wang Q, Jiang H, Xiang X, Deng ZB, Wang B, Zhang L, Roth M, Welti R, Mobley J, Jun Y, Miller D, Zhang HG (2013) Grape exosome-like nanoparticles induce intestinal stem cells and protect mice from DSS-induced colitis. Mol Ther 21(7):1345–1357
Jun I, Kim BR, Park SY, Lee H, Kim J, Kim EK, Seo KY, Kim TI (2020) Interleukin-4 stimulates lipogenesis in meibocytes by activating the STAT6/PPARgamma signaling pathway. Ocul Surf 18(4):575–582
Kahn SE, Hull RL, Utzschneider KM (2006) Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444(7121):840–846
Li Y, Li Z, Ngandiri DA, Llerins Perez M, Wolf A, Wang Y (2022) The molecular brakes of adipose tissue lipolysis. Front Physiol 13:826314
Li M, Chen T, He JJ, Wu JH, Luo JY, Ye RS, Xie MY, Zhang HJ, Zeng B, Liu J, Xi QY, Jiang QY, Sun JJ, Zhang YL (2019a) Plant MIR167e-5p inhibits enterocyte proliferation by targeting beta-catenin. Cells 8(11)
Li M, Chen T, Wang R, Luo JY, He JJ, Ye RS, Xie MY, Xi QY, Jiang QY, Sun JJ, Zhang YL (2019b) Plant MIR156 regulates intestinal growth in mammals by targeting the Wnt/beta-catenin pathway. Am J Physiol Cell Physiol 317(3):C434–C448
Lukasik A, Zielenkiewicz P (2014) In silico identification of plant miRNAs in mammalian breast milk exosomes–a small step forward? PLoS ONE 9(6):e99963
Luo Y, Wang P, Wang X, Wang Y, Mu Z, Li Q, Fu Y, Xiao J, Li G, Ma Y, Gu Y, Jin L, Ma J, Tang Q, Jiang A, Li X, Li M (2017) Detection of dietetically absorbed maize-derived microRNAs in pigs. Sci Rep 7(1):645
Martinez Calejman C, Trefely S, Entwisle SW, Luciano A, Jung SM, Hsiao W, Torres A, Hung CM, Li H, Snyder NW, Villen J, Wellen KE, Guertin DA (2020) mTORC2-AKT signaling to ATP-citrate lyase drives brown adipogenesis and de novo lipogenesis. Nat Commun 11(1):575
Mu J, Zhuang X, Wang Q, Jiang H, Deng ZB, Wang B, Zhang L, Kakar S, Jun Y, Miller D, Zhang HG (2014) Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol Nutr Food Res 58(7):1561–1573
Perry RJ, Peng L, Abulizi A, Kennedy L, Cline GW, Shulman GI (2017) Mechanism for leptin’s acute insulin-independent effect to reverse diabetic ketoacidosis. J Clin Invest 127(2):657–669
Pistor KE, Sepa-Kishi DM, Hung S, Ceddia RB (2015) Lipolysis, lipogenesis, and adiposity are reduced while fatty acid oxidation is increased in visceral and subcutaneous adipocytes of endurance-trained rats. Adipocyte 4(1):22–31
Qi R, Han X, Wang J, Qiu X, Wang Q, Yang F (2021) MicroRNA-489-3p promotes adipogenesis by targeting the Postn gene in 3T3-L1 preadipocytes. Life Sci 278:119620
Scheja L, Heeren J (2019) The endocrine function of adipose tissues in health and cardiometabolic disease. Nat Rev Endocrinol 15(9):507–524
Shen L, Zhang Y, Du J, Chen L, Luo J, Li X, Li M, Tang G, Zhang S, Zhu L (2016) MicroRNA-23a regulates 3T3-L1 adipocyte differentiation. Gene 575(2 Pt 3):761–764
Shen HH, Yang CY, Kung CW, Chen SY, Wu HM, Cheng PY, Lam KK, Lee YM (2019) Raloxifene inhibits adipose tissue inflammation and adipogenesis through Wnt regulation in ovariectomized rats and 3 T3–L1 cells. J Biomed Sci 26(1):62
Smalheiser NR, Torvik VI (2006) Complications in mammalian microRNA target prediction. Methods Mol Biol 342:115–127
Trayhurn P (2005) Endocrine and signalling role of adipose tissue: new perspectives on fat. Acta Physiol Scand 184(4):285–293
Wang S, Cao S, Arhatte M, Li D, Shi Y, Kurz S, Hu J, Wang L, Shao J, Atzberger A, Wang Z, Wang C, Zang W, Fleming I, Wettschureck N, Honore E, Offermanns S (2020) Adipocyte Piezo1 mediates obesogenic adipogenesis through the FGF1/FGFR1 signaling pathway in mice. Nat Commun 11(1):2303
Yamashita T, Lakota K, Taniguchi T, Yoshizaki A, Sato S, Hong W, Zhou X, Sodin-Semrl S, Fang F, Asano Y, Varga J (2018) An orally-active adiponectin receptor agonist mitigates cutaneous fibrosis, inflammation and microvascular pathology in a murine model of systemic sclerosis. Sci Rep 8(1):11843
Yang A, Mottillo EP (2020) Adipocyte lipolysis: from molecular mechanisms of regulation to disease and therapeutics. Biochem J 477(5):985–1008
Zhou LK, Zhou Z, Jiang XM, Zheng Y, Chen X, Fu Z, Xiao G, Zhang CY, Zhang LK, Yi Y (2020a) Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov 6(1):54
Zhou Z, Li X, Liu J, Dong L, Chen Q, Liu J, Kong H, Zhang Q, Qi X, Hou D, Zhang L, Zhang G, Liu Y, Zhang Y, Li J, Wang J, Chen X, Wang H, Zhang J, Chen H, Zen K, Zhang CY (2015) Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res 25(1):39–49
Zhou Z, Zhou Y, Jiang XM, Wang Y, Chen X, Xiao G, Zhang CY, Yi Y, Zhang LK, Li L (2020b) Decreased HD-MIR2911 absorption in human subjects with the SIDT1 polymorphism fails to inhibit SARS-CoV-2 replication. Cell Discov 6:63
Zhu K, Liu M, Fu Z, Zhou Z, Kong Y, Liang H, Lin Z, Luo J, Zheng H, Wan P, Zhang J, Zen K, Chen J, Hu F, Zhang CY, Ren J, Chen X (2017) Plant microRNAs in larval food regulate honeybee caste development. PLoS Genet 13(8):e1006946
Pietruszka A, Jacyno E, KaweCka M, Biel W (2015) The relation between intramuscular fat level in the longissimus muscle and the quality of pig carcasses and meat. Ann Anim Sci 15(4):1031–1041
Acknowledgements
We wish to thank the Key Laboratory of Medical Electronics and Digital Health of Zhejiang Province and the Human Health Situation Awareness of Zhejiang Province for providing the equipment support.
Funding
This study was supported by grants from the Natural Science Foundation of Zhejiang Provincial Program (No. LY22C180005), the Natural Science Foundation of China Program (Nos. 32002244, 31760646, and 32082812), the Natural Science Foundation of Guangdong Provincial Program (No. 2021A1515011310), the Initiated Project of Jiaxing University (No. CD70520012), and the National Innovation and Entrepreneurship Training Program for College Students (No. 202110354032).
Author information
Authors and Affiliations
Contributions
Conceptualization: M.L. and Y.Z.; methodology: F.M. and J.Z.; software: Y.P.; validation: Q.X., and J.S.; data analysis: T.C.; data curation: T.C.; writing/preparation of the original draft: M.L. and F.M.; writing with review and editing: M.L. and T.C.; visualization: F.M. and Y.P.; supervision: R.S.; project administration: R.S. and M.L.; funding acquisition: Y.Z. and M.L.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Chen, T., Ma, F., Peng, Y. et al. Plant miR167e-5p promotes 3T3-L1 adipocyte adipogenesis by targeting β-catenin. In Vitro Cell.Dev.Biol.-Animal 58, 471–479 (2022). https://doi.org/10.1007/s11626-022-00702-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-022-00702-w