Abstract
Our studies on lignicolous aquatic fungi in Thailand, Sweden, and the UK resulted in the collection of three new Halobyssothecium species (H. bambusicola, H. phragmitis, H. versicolor) assigned to Lentitheciaceae (Pleosporales, Dothideomycetes). Multi-loci phylogenetic analyses of the combined large subunit, small subunit, internal transcribed spacers of ribosomal DNA, and the translation elongation factor 1-alpha sequence data enabled a revision of the taxa assigned to Lentithecium and the transfer of L. cangshanense, L. carbonneanum, L. kunmingense, L. unicellulare, and L. voraginesporum to Halobyssothecium. Collection of an asexual morph of L. lineare and phylogenetic analysis confirmed its taxonomic placement in Keissleriella. Detailed descriptions and illustrations of H. bambusicola, H. phragmitis, and H. versicolor are provided.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pleosporales, typified by Pleospora herbarum (Pers.) Rabenh. (Pleosporaceae), was formally established by Luttrell and Barr (in Barr 1987) and characterized by perithecioid ascomata, usually with a papillate apex, ostiolate, cellular pseudoparaphyses, and bitunicate asci. Phylogenetic studies of Pleosporales have been provided by Schoch et al. (2009), Zhang et al. (2009a, 2012), Hyde et al. (2013), Liu et al. (2017), and Hongsanan et al. (2020). Lumbsch and Huhndorf (2010) included 28 families and 175 genera in Pleosporales, with 12 genera listed under Pleosporales, genera incertae sedis. Hyde et al. (2013) accepted 88 families in Pleosporales. Wijayawardene et al. (2020) and Hongsanan et al. (2020) included 91 families in Pleosporales. Ecologically, the order includes saprotrophs, parasites, pathogens, epiphytes, and endophytes (Hongsanan et al. 2020).
Zhang et al. (2009b) established Lentitheciaceae with Lentithecium fluviatile (Aptroot & Van Ryck.) K.D. Hyde, J. Fourn. & Ying Zhang as the genus and species type, and included L. arundinaceum (Sowerby) K.D. Hyde, J. Fourn. & Ying Zhang, L. aquaticum Ying Zhang, J. Fourn. & K.D. Hyde, Stagonospora macropycnidia Cunnell, Wettsteinina lacustris (Fuckel) Shoemaker & C.E. Babc., Keissleriella cladophila (Niessl) Corbaz, and Katumotoa bambusicola Kaz. Tanaka & Y. Harada. Suetrong et al. (2009) also referred Massarina phragmiticola Poon & K.D. Hyde to the new family. Lentitheciaceous taxa are saprobic on herbaceous and woody plants having narrow peridia, fusiform to broadly cylindrical pseudoparaphyses, hyaline ascospores with 1–3-transverse septa and containing refractive globules, surrounded by a mucilaginous sheath or extended appendage-like sheaths and asexual morphs producing stagonospora-like or dendrophoma-like asexual morphs (Zhang et al. 2012; Hyde et al. 2013; Wanasinghe et al. 2014). Fourteen genera from different habitats are included in Lentitheciaceae based on molecular data: Darksidea (Knapp et al. 2015), Halobyssothecium (Dayarathne et al. 2018), Katumotoa (Tanaka and Harada 2005), Keissleriella (Höhnel 1919), Lentithecium (Zhang et al. 2009b), Murilentithecium (Wanasinghe et al. 2014), Neoophiosphaerella (Tanaka et al. 2015), Phragmocamarosporium (Wijayawardene et al. 2015), Pleurophoma (de Gruyter et al. 2009; Crous et al. 2015), Poaceascoma (Phookamsak et al. 2015), Pseudomurilentithecium (Hyde et al. 2020b), Setoseptoria (Quaedvlieg et al. 2013), Tingoldiago (Hirayama et al. 2010), and Towyspora (Li et al. 2016).
Lentithecium was proposed to accommodate Massarina arundinacea (Sowerby) Leuchtm., M. fluviatilis Aptroot & Van Ryck., and Keissleriella linearis E. Müll. ex Dennis (Zhang et al. 2009b). The genus currently contains ten species that were described from aquatic habitats, seven from freshwater, and three from marine environments. Lentithecium species have been described from submerged wood (Tanaka et al. 2005, 2015; Hyde et al. 2016; Su et al. 2016; Crous et al. 2018) and submerged parts of plant host species (Juncus, Phragmites, Fraxinus, Alnus, and Platanus) (Kohlmeyer et al. 1996; Van Ryckegem and Aptroot 2001; Suetrong et al. 2009; Zhang et al. 2009b). Lentithecium is characterized by its immersed to semi-immersed, globose to subglobose ascomata, a thin peridium, cellular pseudoparaphyses, short pedicellate asci and fusoid or filiform, subglobose, hyaline, brown, uni- to multi-septate ascospores, usually surrounded by a sheath (Zhang et al. 2009b; Hyde et al. 2013, 2016).
Halobyssothecium was introduced by Dayarathne et al. (2018) to accommodate several taxa variously described under Pleospora obiones P. Crouan & H. Crouan by Crouan and Crouan (1867) and Leptosphaeria discors Sacc. & Ellis by Saccardo (1882). This “taxon” had been assigned to various genera: Metasphaeria (Saccardo 1883), Heptameria (Cooke 1889), and Passeriniella (Apinis and Chesters 1964; Hyde and Mouzouras 1988; Khashnobish and Shearer 1996). Various studies have shown that Pleospora obiones/Leptosphaeria discors are synonyms, but clearly do not belong in any of these genera (Khashnobish and Shearer 1996). Jones (1962), Cavaliere (1968), and Webber (1970) reported Leptosphaeria discors collections with larger ascospores than those by Crouan and Crouan (1867) indicating that there might be a second morphologically similar species. Halobyssothecium obiones (P. Crouan & H. Crouan) Dayarathne, E.B.G. Jones & K.D. Hyde has a worldwide distribution in temperate regions and occurs as a saprobe of Agropyron junceiforme, Halimione portulacoides, Spartina species, on intertidal wood, bamboo, and exposed test panels of Betula pubescens and Fagus sylvatica (Kohlmeyer and Kohlmeyer 1979; Jones et al. 2019). Devadatha et al. (2020) introduced Halobyssothecium estuariae B. Devadatha, Calabon, K.D. Hyde & E.B.G. Jones collected on a dead culm of Phragmites communis from Slebech Estuary, Pembrokeshire, UK, which resembles the generic type H. obiones in possessing subglobose or ellipsoidal, carbonaceous ascomata, conical papilla and ascospores with brown central cells and hyaline end cells (Dayarathne et al. 2018). However, H. estuariae is distinct from H. obiones in having a longer and narrower papilla (65–85 × 55–85 vs. 25–35 × 130–145 μm) and smaller ascospores (20–44 × 4–9 vs. 28–47 × 10–18 μm). Halobyssothecium obiones and H. estuariae differ by 5.1% (22/431 bp) in ITS and 3.12% (28/895 bp) in TEF1-α sequence data.
Keissleriella, typified by K. aesculi (Höhn.) Höhn., is characterized by an ostiolar neck covered by short dark setae (Tanaka et al. 2015; Hongsanan et al. 2020). Keissleriella is the most speciose genus in Lentitheciaceae with 46 epithets listed in Species Fungorum (http://www.speciesfungorum.org/Names/Names.asp; accessed on December 2020) and 38 morphological species, 25 of which have molecular data. Sequence data for the type species of Keissleriella is unavailable, but phylogenetic studies confirmed its placement within Lentitheciaceae (Tanaka et al. 2015; Tibpromma et al. 2017; Hongsanan et al. 2020).
In the present study, a phylogenetic tree of taxa in Lentitheciaceae was constructed based on sequence data of four loci (LSU, SSU, ITS, TEF1-α) to reevaluate the taxonomic status of Halobyssothecium and Lentithecium. The latest treatments and updated accounts of Lentitheciaceae in Dayarathne et al. (2018), Hongsanan et al. (2020), and Wijayawardene et al. (2020) are followed in this paper. The insights from the multi-loci analyses and morphological observations reveal three new species of Halobyssothecium and confirm the taxonomic placement of Lentithecium lineare (E. Müll. ex Dennis) K.D. Hyde, J. Fourn. & Ying Zhang in Keissleriella, and L. cangshanense Z.L. Luo, X.J. Su & K.D. Hyde, L. carbonneanum J. Fourn., Raja & Oberlies, L. kunmingense Dong, H. Zhang & K.D. Hyde, L. unicellulare Abdel-Aziz and L. voraginesporum Abdel-Wahab, Bahkali & E.B.G. Jones in Halobyssothecium. The transfers are made, and descriptions, photographic plates, and multi-loci phylogenetic analyses are provided.
Materials and methods
Sample collection, morphological observation, and fungal isolation
Samples of submerged decayed wood were collected from a freshwater stream in Chiang Mai, Thailand. Dead and decaying Halimione portulacoides was collected from Hayling Island bridge, Hampshire, UK. Drift culms and stems of Phragmites sp. were obtained from Sudersand and Kappelshamnsviken in Gotland, Sweden. The samples were observed using a stereomicroscope for the presence of fruiting bodies. Micromorphological features were photographed using a Motic SMZ 168 Series dissection microscope for fungal structures on the woody substrate while microscopic characters were documented using a Nikon Eclipse 80i microscope. Single spore isolation was used to obtain pure cultures and colonial characteristics described. Herbarium-type specimens are deposited in Mae Fah Luang University (MFLU). Ex-type and ex-paratype living cultures are deposited at Mae Fah Luang University Culture Collection (MFLUCC). The new species and combinations were registered in Faces of Fungi (http://www.facesoffungi.org/; Jayasiri et al. 2015) and Index Fungorum database (http://www.indexfungorum.org/names/IndexFungorumRegisterName.asp).
DNA extraction, PCR amplification, and sequencing
Fungal mycelia from pure cultures grown in malt extract agar (MEA) for 30 days were scraped using a sterilized scalpel and kept in a sterilized 1.5 mL microcentrifuge tube. Genomic DNA was extracted using the Biospin Fungus Genomic DNA Extraction Kit (BioFlux®, China) following the manufacturer’s protocol. Polymerase chain reaction (PCR) was used to amplify four markers: the large subunit (LSU), small subunit (SSU), internal transcribed spacers (ITS) of rDNA, and the translation elongation factor 1-alpha gene (TEF1-α). The LSU was amplified using the primers LR0R and LR5 (Vilgalys and Hester 1990). The SSU was amplified using the primers NS1 and NS4 (White et al. 1990). For ITS, primers ITS5 and ITS4 were used (White et al., 1990). TEF1-α was amplified using primers EF1-983F and EF1-2218R (Rehner 2001). Polymerase chain reaction was performed in a volume of 25 μl, which contained 12.5 μl of 2× Power Taq PCR Master Mix (Bioteke Co., China), 1 μl of each primer (10 pM), 1 μl genomic DNA, and 9.5 μl double-distilled water (ddH2O). The PCR thermal cycle program for LSU, SSU, ITS, and TEF1-α amplification were as follows: initial denaturing step of 94 °C for 3 min, followed by 40 cycles of denaturation at 94 °C for 45 seconds, annealing at 56 °C for 50 seconds, elongation at 72 °C for 1 min, and final extension at 72 °C for 10 min. Agarose gel electrophoresis was done to confirm the presence of amplicons at the expected molecular weight. PCR products were purified and sequenced with the primers mentioned above at a commercial sequencing provider (Beijing Qingke Biotechnology Co., Ltd). A BLASTn search of the newly generated sequences was carried out to exclude contamination and to search for related taxa in GenBank database (www.ncbi.nlm.nih.gov/blast/).
Phylogenetic analyses
The taxa table was assembled based on the closest matches from the BLASTn search results and from recently published data in Dayarathne et al. (2018) and Devadatha et al. (2020). Sequences generated from the four markers were analyzed along with other sequences retrieved from GenBank (Table 1). Four datasets, one for each marker, were aligned with MAFFT v. 7 using the web server (http://mafft.cbrc.jp/alignment/server; Katoh et al. 2019) with the following settings: L-INS-i tree-based iterative refinement methods, 20PAM/k = 2 scoring matrix for nucleotide sequences and 1.53 gap opening penalty. Alignment was further refined manually, where necessary, using BioEdit v.7.0.9.0 (Hall 1999). Aligned sequences were automatically trimmed using TrimAl v. 1.3 on the web server (http://phylemon.bioinfo.cipf.es/utilities.html). The online tool “ALTER” (Glez-Peña et al. 2010) was used to convert the alignment file to phylip and nexus formats. Phylogenetic analyses of both individual and combined gene data were performed using maximum likelihood (ML), maximum parsimony (MP), and Bayesian inference (BI).
Maximum parsimony (MP) analysis was performed using the heuristic search option with 1000 random taxa addition and tree bisection and reconnection (TBR) as the branch-swapping algorithm in PAUP* 4.0b4 (Swofford 2002). All characters were unordered and of equal weight and gaps were treated as missing data. Maxtrees were unlimited, branches of zero length were collapsed and all multiple, equally parsimonious trees were saved. Clade stability was assessed using a bootstrap (BS) analysis with 1000 replicates, each with ten replicates of random stepwise addition of taxa (Hillis and Bull 1993). Descriptive tree statistics for parsimony (tree length [TL], consistency index [CI], retention index [RI], relative consistency index [RC], and homoplasy index [HI]) were calculated for trees generated under different optimality criteria.
Maximum likelihood analysis was performed using RAxML-HPC2 on XSEDE on the CIPRES web portal (Stamatakis 2006, 2014; Stamatakis et al. 2008) (http://www.phylo.org/portal2/; Miller et al. 2010). The GTR+GAMMA model of nucleotide evolution was used. RAxML rapid bootstrapping of 1,000 replicates was performed. The best-fit evolutionary models for individual and combined datasets were estimated under the Akaike Information Criterion (AIC) using jModeltest 2.1.10 on the CIPRES web portal and each resulted to the GTR+I+G model (Nylander 2004; Darriba et al. 2012). Bayesian inference analyses were performed using MrBayes v. 3.2.6 on XSEDE at the CIPRES webportal (Ronquist and Huelsenbeck 2003), using the parameter setting of two parallel runs, four chains, the run for 4,000,000 generations at which point the standard deviation of split frequencies was below 0.01. Trees were sampled every 1,000 generations and all other parameters were left as default. Bayesian analysis resulted in 4,000 trees after the run wherein the first 1,000 trees, 25% of the total, were in the burn-in phase and were discarded. The remaining 3,000 trees were used to calculate the posterior probability (PP). Newly generated sequences have been deposited in GenBank (Table 1).
Genealogical concordance phylogenetic species recognition analysis
New species and their most closely related species were analyzed using the Genealogical concordance phylogenetic species recognition (GCPSR) model. A pairwise homoplasy index (PHI) (Bruen et al. 2006) test was performed in SplitsTree4 (Huson 1998; Huson and Bryant 2006) as described by Quaedvlieg et al. (2014). This was done to determine the recombination level within phylogenetically closely related species using a four-locus concatenated dataset for new species of Halobyssothecium. The test detects incompatibility between pairs of sites regarding whether there is genealogical history that can be inferred parsimoniously that does not involve any recurrent or convergent mutations. Pairwise homoplasy index below a 0.05 threshold (Фw < 0.05) indicates that there is significant recombination present in the dataset. The relationships between closely related species were visualized by constructing a split graph, using both the LogDet transformation and splits decomposition options.
Results
Phylogenetic analyses
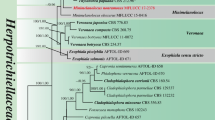
The combined LSU, SSU, ITS and TEF1-α dataset comprised of 133 taxa from Lentitheciaceae, with Corynespora cassiicola (Berk. & M.A. Curtis) C.T. Wei (CBS 100822) and C. smithii (Berk. & Broome) M.B. Ellis (CABI5649b) as outgroup taxa (Table 1). The analyzed dataset, after trimming, comprised a total 3,578 characters including gaps (LSU = 1,274 bp, SSU = 916 bp, ITS = 473 bp, TEF1-α = 915 bp) with 1,632 distinct alignment patterns and 28.64% proportion of gaps and completely undetermined characters, 2,235 constant, 414 parsimony uninformative and 940 parsimony informative characters. The MP analysis resulted a single most parsimonious tree (TL = 5,457, CI = 0.364, RI = 0.674, RC = 0.245, HI = 0.636). The ML analysis for the combined dataset provided the best scoring tree (Fig. 1) with a final ML optimization likelihood value of -32434.024914 (ln). Parameters for the GTR+I+G model of the combined LSU, SSU, ITS and TEF1-α dataset are as follows: estimated base frequencies; A = 0.241074, C = 0.248510, G = 0.273533, T = 0.236882; substitution rates AC = 1.038579, AG = 2.219296, AT = 1.397250, CG = 1.151737, CT = 6.450277, GT = 1.000000; gamma distribution shape parameter α = 0.228421. The Bayesian analysis indicated the average standard deviation of split frequencies at the end of total MCMC generations is 0.007035. Phylogenetic analyses of the combined data matrix resulted in well-resolved clades (Fig. 1). The tree topologies resulted from maximum likelihood (ML), maximum parsimony (MP), and Bayesian posterior probabilities (BYPP) analyses were congruent.
Phylogenetic tree generated from maximum likelihood (ML) analysis based on LSU, SSU, ITS and TEF1-α sequence data for the species from Lentitheciaceae and closely related families in Pleosporales. Bootstrap support values for maximum likelihood (ML) and maximum parsimony (MP) higher than 50% and Bayesian posterior probabilities (BYPP) greater than 0.90 are indicated above the nodes in this order. The new isolates are represented in blue. The ex-type strains are indicated in bold. The tree is rooted to Corynespora cassiicola (CBS 100822) and C. smithii (CABI5649b) (Corynesporascaceae). Bar = 0.04 estimated number of nucleotide substitutions per site per branch
In the phylogenetic analysis (Fig. 1), Halobyssothecium formed a well-supported monophyletic clade, separate from Lentithecium (99% ML, 95% MP, 1.00 BYPP). Three novel Halobyssothecium species, H. bambusicola, H. phragmitis and H. versicolor grouped with the other Halobyssothecium species in Lentitheciaceae. Moreover, five species of Lentithecium (L. cangshanense, L. carbonneanum, L. kunmingense, L. unicellulare, L. voraginesporum) clustered with Halobyssothecium. Therefore, these five Lentithecium species are transferred to Halobyssothecium in this study. Halobyssothecium bambusicola MFLUCC 20–0226 and H. kunmingense KUMCC 19–0101 were strongly supported as sister species (100% ML, 100% MP, 1.00 BYPP) and clustered with H. phragmitis (MFLUCC 20–0223, MFLUCC 20–0225) with high support (93% ML, 80% MP, 1.00 BYPP). Halobyssothecium versicolor MFLUCC 20–0222 forms a distinct lineage and basal to other Halobyssothecium species. Lentithecium clioninum (Kaz. Tanaka, Sat. Hatak. & Y. Harada) Kaz. Tanaka & K. Hiray. and L. pseudoclioninum Kaz. Tanaka & K. Hiray. clustered together with L. fluviatile, the type species of Lentithecium (99% ML, 96% MP, 1.00 BYPP). Furthermore, L. lineare MFLUCC 20–0224 clustered with the other two strains of L. lineare (IFRD2008, MFLUCC 19–0410) (100% ML, 100% MP, 1.00 BYPP).
The relationships between the three new species of Halobyssothecium were visualized by constructing a split graph and PHI-test revealed significant genetic recombination levels between two strains of H. phragmitis suggesting that they are conspecific. The presence of recombination among fungal isolates is the hallmark that these belong to the same biological species. No significant recombination events were observed between H. bambusicola, H. kunmingense, and H. phragmitis indicating that these are different species (Fig. 2). PHI-test returns the probability of observing the data under the null hypothesis of no recombination.
Taxonomy
Halobyssothecium Dayar., E.B.G. Jones & K.D. Hyde
Saprobic on salt marsh halophytes and submerged decaying wood in aquatic habitats. Sexual morph: Ascomata immersed, semi-immersed or erumpent, scattered to clustered, globose to subglobose or ellipsoidal, carbonaceous, dark brown to black, gregarious, ostiolate. Peridium comprising of only pseudoparenchyma or two layers: outer layer of brown, inner layer of elongated, hyaline cells. Pseudoparaphyses cellular, septate, branched. Asci 8-spored, bitunicate, fissitunicate, cylindric-clavate to subcylindrical, short pedicellate, thick-walled, with or without an ocular chamber. Ascospores overlapping uni- to bi-seriate, clavate, ellipsoid, subcylindrical ovoid or fusoid with rounded ends, versicolored, initially hyaline when young to pale brown, golden brown or brown when mature, end cells hyaline, central cells brown, 1–3-septate, constricted at the septa, guttulate, slightly curved, lacking gelatinous sheath or appendages, slimy material without well-defined sheath. Asexual morph: Coelomycetous. Conidiomata pycnidial, immersed, erumpent at maturity, solitary or aggregated, unilocular, globose to subglobose, ellipsoidal, dark brown to black, ostiolate. Ostiole single, circular to subcylindrical, papillate, dark brown to black, centrally located. Conidiomatal wall composed of thick-walled, dark brown cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells enteroblastic, phialidic, determinate, smooth-walled, hyaline, aseptate, globose to subglobose, ellipsoidal, cylindrical to subcylindrical. Conidia spherical to globose, subglobose, ovate to obovate, ellipsoidal, clavate to subclavate, lageniform, hyaline, aseptate, straight to slightly curved, guttulate, smooth, and thick-walled. Chlamydospores apical, rarely intercalary, single or in chains, branching, filamentous, filiform to narrowly fusiform straight or curved, catenate, rarely solitary, branched, septate, with thickened septa, brown to dark brown at the septa, smooth-walled.
Type species: Halobyssothecium obiones (P. Crouan & H. Crouan) Dayar., E.B.G. Jones & K.D. Hyde, Mycological Progress 17 (10): 1165 (2018)
Notes: Two species were included in Halobyssothecium, H. obiones and H. estuariae (Dayarathne et al. 2018; Devadatha et al. 2020), collected from various host substrates in temperate regions. In the present study, three collections of morphologically distinct isolates were encountered, two were asexual morphs (H. bambusicola and H. phragmitis) and one sexual morph (H. versicolor), which advances the current understanding of how complex the genus is. The complexity was noted by Devadatha et al. (2020) based on previous collections by various authors. For instance, two morphologically similar taxa of H. obiones were collected but differed in ascospore measurements (24–38 × 8–14 μm vs. 38–56 × 16–22 μm) (Jones 1962; Cavaliere 1968; Webber 1970), but no sequence data was available at that time to distinguish them. Halobyssothecium versicolor agrees with the generic description of the genus and its placement in the phylogenetic tree redefines what comprises Halobyssothecium. Currently, the Lentithecium clade includes L. fluviatile, L. clioninum and L. pseudoclioninum, while L. cangshanense, L. carbonneanum, L. kunmingense, L. unicellulare, and L. voraginesporum grouped within the Halobyssothecium clade and are transferred herein.
Halobyssothecium bambusicola M.S. Calabon, Boonmee, E.B.G. Jones & K.D. Hyde, sp. nov. (Fig. 3)
Index Fungorum number: IF558089; Facesoffungi number: FoF 09430
Halobyssothecium bambusicola (MFLU 20–0549, holotype). a Host. b–d Appearance of conidiomata on host surface releasing conidia in a cirrus (arrow). e Vertical section of conidioma. f Conidiomatal wall. g–j Developing conidia attach to conidiogenous cell. k–r Conidia. s–t Germinated conidia. u Colony on MEA (obverse, reverse). Scale bars: a = 200 mm; b = 1 mm; c–e = 500 μm; f = 50 μm; g–t = 10 μm
Etymology: the specific epithet “bambusicola” refers to the host, of which the fungus was collected
Holotype: MFLU 20–0549
Saprobic on decaying bamboo culms submerged in freshwater habitat. Sexual morph: Undetermined. Asexual morph: Conidiomata 350–470 μm high, 230–260 μm wide (x̅ = 415.4 × 238.6, n = 10), pycnidial, immersed, erumpent at maturity, solitary or aggregated, globose, unilocular, dark brown to black, ostiolate. Ostiole 150–160 × 170–180 μm, single, circular to subcylindrical, centrally located. Conidiomatal wall 14–28 μm, composed of thick-walled, dark brown cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 6–45 × 2–5 μm (x̅ = 19.7 × 3.3, n = 30), enteroblastic, phialidic, hyaline, aseptate, cylindrical to subcylindrical. Conidia 6–12 × 5–10 μm (x̅ = 8.7 × 6.8, n = 50), spherical to globose, obovate, ellipsoidal, subclavate, hyaline, aseptate, guttulate, smooth, and thick-walled.
Culture characteristics: On MEA, colony circular with filamentous margin, reaching 25–30 mm diam. in 25 days at 25 °C, brown to grayish brown from above, yellowish brown to dark brown from below, surface rough, dry, raised, with dense mycelia, edge filiform.
Material examined: THAILAND, Chiang Mai Province, on submerged bamboo culm in a stream, 11 February 2019, M.S. Calabon (MFLU 20–0549, holotype), ex-type living culture MFLUCC 20–0226
Notes: Several species of freshwater fungi growing on submerged bamboo have been recorded, e.g. Acrodictys liputii L. Cai, K.Q. Zhang, McKenzie, W.H. Ho & K.D. Hyde, Annulatascus liputii L. Cai & K.D. Hyde, Ascoyunnania aquatica L. Cai & K.D. Hyde, Cataractispora receptaculorum W.H. Ho, K.D. Hyde & Hodgkiss, Dictyosporella thailandensis W. Dong, H. Zhang & K.D. Hyde, Fluminicola saprophytica W. Dong, H. Zhang & K.D. Hyde, Dictyochaeta curvispora L. Cai, McKenzie & K.D. Hyde, D. plovercovensis Goh & K.D. Hyde, Diluviicola aquatica W. Dong, H. Zhang & K.D. Hyde, Linocarpon bambusicola L. Cai & K.D. Hyde, Ophioceras guttulatum C.K.M. Tsui, H.Y.M. Leung, K.D. Hyde & Hodgkiss, Microthecium sepedonioides (Preuss) Y. Marín, Stchigel, Guarro & Cano, Payosphaeria minuta H.Y.M. Leung, Pseudoproboscispora thailandensis W. Dong, H. Zhang & K.D. Hyde, and Saccardoella minuta L. Cai & K.D. Hyde (Cai et al. 2002a, b, 2003, 2004, 2005, 2006; Ho et al. 2004; Zhang et al. 2017).
Halobyssothecium bambusicola closely resembles H. kunmingense, and H. unicellulare. Halobyssothecium kunmingense has wider conidiomata (210–250 × 320–350 μm vs. 350–470 × 230–260 μm), a thicker peridium (60–80 μm vs 14–28 μm), smaller conidiogenous cells (5–19 × 2–5 μm vs. 6–45 × 2–5 μm) and larger globose to obovoidal conidia (8–14 × 5–8 μm vs 6–12 × 5–10) with large guttules, compared to H. bambusicola (Dong et al. 2020). The guidelines on species delimitation for new species by Jeewon and Hyde (2016) was followed and pairwise comparison of ribosomal ITS sequences showed 15 nucleotide base pair differences among the 800 nucleotides analyzed between H. bambusicola and H. kunmingense. Moreover, a SplitsTree analysis supports the introduction of H. bambusicola (Fig. 2).
Halobyssothecium bambusicola has larger conidiomata (350–470 μm high × 230–260 μm wide vs. 115–235 μm high × 140–235 μm wide) and larger globose to obovoidal conidia (6–12 × 5–8 μm vs. 6–9 × 4–5 μm) with large guttules as compared to H. unicellulare (Hyde et al. 2016). Halobyssothecium bambusicola differs from H. phragmitis in conidial shape (globose to obovate conidia vs. ovoid to fusoid-ellipsoidal). Halobyssothecium bambusicola and H. phragmitis differ by 6.84% (36/526 bp) and 2.63% (25/952 bp) in ITS and TEF1-α, respectively. The multigene phylogenetic analysis placed H. bambusicola within Halobyssothecium in a well-supported subclade with H. kunmingense (100% ML, 100% MP, 1.00 BYPP) and the coelomycetous marine H. phragmitis (93% ML, 80% MP, 1.00 BYPP).
Halobyssothecium phragmitis M.S. Calabon, E.B.G. Jones, S. Tibell & K.D. Hyde, sp. nov. (Fig. 4)
Index Fungorum number: IF558090; Facesoffungi number: FoF 09431
Halobyssothecium phragmitis (MFLU 20–0550, holotype). a Host. b Appearance of conidiomata on host surface. c Vertical section of conidioma. d Ostiole. e Conidiomatal wall. f–j Developing conidia attach to conidiogenous cells. k–r Conidia. s Germinated conidium. t–u Colony on MEA: from t obverse, u reverse. Scale bars: b, c = 200 μm, d = 100 μm, e–s = 10 μm
Etymology: In reference to the host genus Phragmites, from which the species was isolated.
Holotype: MFLU 20–0550
Saprobic on dead Phragmites culm and stem. Sexual morph: Undetermined. Asexual morph: Conidiomata 205–340 μm high, 215–280 μm wide, solitary, scattered, immersed to slightly immersed, pycnidial, subglobose to ellipsoidal, unilocular, black, with indistinct ostioles. Ostioles 82–96 μm, central, circular, papillate, dark brown to black. Conidiomatal wall 13.3–31 μm, thick-walled, 7–9 layers, comprising of dark brown cells, of textura angularis, inner layer comprising hyaline gelatinous layer, thickening at the upper and basal zone. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 8–18 × 1–5 μm (x̅ = 11.6 × 3.2 μm, n = 20), enteroblastic, phialidic, cylindrical to lageniform, determinate, hyaline, formed from inner layers of conidiomata. Conidia 9–19 × 2–6 μm (x̅ = 13.7–4.1 μm, n = 50), cylindrical, fusoid-ellipsoidal, straight or slightly curved, hyaline, aseptate to 1–2-septate, unicellular, mostly with one large central guttule per cell, smooth-walled.
Culture characteristics: Conidia germinated on MEA within 24 h. Colonies on MEA, reaching 10–12 mm diam. in 14 days at 25 °C. Mycelium superficial, white, flattened, hairy, dense, circular, flattened, margin entire; reverse pale brown.
Material examined: SWEDEN, Gotland, Kappelshamnsviken, on dead Phragmites culm (Poaceae), 7 March 2019, E.B.G. Jones, GJ653 (MFLU 20–0550, holotype), ex-type living cultures MFLUCC 20–0223; ibid, Sudersand, on dead Phragmites (Poaceae) stem, 7 March 2019, E.B.G. Jones, GJ659 (MFLU 20–0552, paratype), ex-paratype living culture MFLUCC 20–0225.
Notes: Halobyssothecium phragmitis resembles Stagonospora macropycnidia but the former species has smaller conidiomata (205–340 μm high × 215–280 μm wide vs. 410–1020 μm high × 120–380 μm wide), and smaller conidia (9–19 × 2–6 μm vs. 22–42 × 2.5–5 μm) (Cunnell 1961). Setoseptoria phragmitis Quaedvl., Verkley & Crous is distinct from H. phragmitis with smaller conidiomata (up to 200 μm vs. 205–340 μm) and longer subcylindrical conidia (19–38 × 3.5–4 μm vs. 9–19 × 2–6 μm) (Quaedvlieg et al. 2013). Phragmocamarosporium platani Wijayaw., Yong Wang bis & K.D. Hyde differs from H. phragmitis with smaller conidiomata (100–320 μm high, 150–300 μm diam. vs. 205–340 μm high, 215–280 μm wide) and larger brown conspicuous phragmospores (12–13 × 5–7.5 μm vs. 9–19 × 2–6 μm) (Wijayawardene et al. 2015). Pleurophoma ossicola Crous, Krawczynski & H.-G. Wagner differs from H. phragmitis with smaller conidia (3–5 × 1.5–2 μm vs. 9–19 × 2–6 μm) (Crous et al. 2015). Murilentithecium clematidis Wanas., Camporesi, E.B.G. Jones & K.D. Hyde is distinct from H. phragmitis with larger conidiomata (0.5–1.5 mm diam vs. 205–340 μm) (Wanasinghe et al. 2014). Keissleriella quadriseptata Kaz. Tanaka & K. Hiray. differs from H. phragmitis with larger cylindrical conidia (25–32 × 6–8.5 μm vs. 9–19 × 2–6 μm) (Tanaka et al. 2015). Based on multi-loci phylogenetic analyses, the above mentioned species are phylogenetically distinct to H. phragmitis.
Halobyssothecium phragmitis is phylogenetically close to H. bambusicola and H. kunmingense (93% ML, 80% MP, 1.00 BYPP). It differs from the latter with ovoidal to fusoid-ellipsoidal conidia. Halobyssothecium kunmingense has 14 base pair differences (800 bp, 1.75%) with H. bambusicola in ITS region.
Halobyssothecium versicolor M.S. Calabon, E.B.G. Jones & K.D. Hyde, sp. nov. (Fig. 5)
Index Fungorum number: IF558091; Facesoffungi number: FoF 09432
Halobyssothecium versicolor (MFLU 19–0676, holotype). a Host. b–e Appearance of ascomata on the host. f–h Sections of ascomata. i Ostiole. j Section of peridium. k Pseudoparaphyses. l–n Asci. o Apex of ascus. p Colony on MEA. q–w Ascospores. x Germinated conidium. Scale bar: a = 20 mm, b–e = 500 μm, f–h = 200 μm, i–n = 50 μm, o, q–x = 20 μm
Etymology: Referring to the versicolored ascospore
Holotype: MFLU 19–0676
Saprobic on Halimione portulacoides in intertidal habitat. Sexual morph: Ascomata 265–510 μm high, 365–530 μm wide (x̅ = 408 × 459, n = 10), superficial to semi-immersed, clustered, sometimes solitary, scattered, subglobose or ellipsoidal, dark brown to black, carbonaceous, conspicuous at the surface, uni- to bi-loculate, ostiolate, with periphyses. Ostiolar neck 105–190 μm long, 95–175 μm wide (x̅ = 150 × 135, n = 10) central, papillate, rounded, short, crest-like, dark brown, composed of several layers of pseudoparenchymatous cells. Peridium 37–94 μm thick, comprising two layers: outer layer of brown pseudoparenchyma; inner layer of elongated, hyaline cells. Pseudoparaphyses 2–3 μm wide, septate, hyaline, filiform, branched and anastomosing above the asci. Asci 137–173 × 17–12 μm (x̅ = 153.4 × 14.7 μm, n = 20), 8-spored, clavate to subcylindrical, short pedicellate with an ocular chamber. Ascospores 18–41 × 6–12 μm (x̅ = 27.4 × 8.6, n = 20), overlapping, uniseriate to biseriately arranged, versicolored, central cells are pale brown to dark brown, end cells hyaline, 1-septate at an early stage, 3-septate when mature, and constricted at the septa, slightly curved, lacking gelatinous sheaths or appendages. Asexual morph: Undetermined.
Culture characteristics: Ascospores germinated on MEA within 24 h. Colonies on MEA, reaching 10–15 mm diam. in 15 days at 25 °C. Mycelium superficial, initially pale yellow, becoming yellowish brown with age, hairy, effuse with wavy edge, dense, circular, raised, undulate, reverse dark yellowish brown.
Material examined: UK, Hampshire, Hayling Island bridge, on dead Halimione portulacoides (Amaranthaceae), 28 February 2019, E.B.G Jones, GJ597 (MFLU 19–0676, holotype), ex-type living cultures MFLUCC 20–0222.
Notes: Halobyssothecium versicolor resembles H. obiones and H. estuariae in having versicolored ascospores with brown central cells and hyaline end cells. Halobyssothecium versicolor differs from H. obiones with larger ascomata (265–510 μm high, 365–530 μm diam. vs. 360–400 μm high, 340–380 μm diam.) and smaller ascospores (18–41 × 6–12 μm vs. 28–47 × 10–18 μm) (Dayarathne et al. 2018). The asexual morph was not observed in the culture but Halobyssothecium species have xylomyces-like chlamydospores (Devadatha et al. 2020) and phoma-like conidia (Kohlmeyer and Kohlmeyer 1979; Calado et al. 2015).
Phylogenetic analysis shows that Halobyssothecium versicolor clustered within Lentitheciaceae and basal to other Halobyssothecium species. Halobyssothecium versicolor is phylogenetically close to H. bambusicola, H. kunmingense, and H. phragmitis. A comparison of ITS and TEF1-α sequence data of H. versicolor differs by 40 (8.97%, 446 bp) and 56 (6.26%, 895 bp) base pairs with H. obiones, type species of the genus.
Keissleriella linearis E. Müll. ex Dennis, Kew Bulletin 19 (1): 120 (1964)
Facesoffungi number: FoF 09433
Saprobic on decaying culm of Phragmites sp. Sexual morph: [for descriptions and illustrations, see Dennis (1964)]. Asexual morph: Conidiomata 42.6–53 μm high, 15.5–22.2 μm wide (x̅ = 51 × 18.8 μm, n = 10), black, solitary, scattered, immersed, pycnidial, subglobose to ellipsoidal, unilocular. Conidiomatal wall 2.2–7.4 μm, thick-walled, 7–9 layers, comprising of dark brown cells, of textura angularis to textura globosa, inner layer comprising hyaline gelatinous layer, thickening at the upper zone. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 4.1–7.7 × 1.6–3.8 μm, (x̅ = 5.8 × 3.1 μm, n = 20), enteroblastic, phialidic, cylindrical to lageniform, determinate, hyaline. Conidia 4.0–6.5 × 1.0–2.5 μm (x̅ = 5.4 × 1.8 μm, n = 50), ovoid, obovoidal, cylindrical, fusoid-ellipsoidal, straight, or slightly curved, unicellular, hyaline, guttulate, smooth-walled. Beta conidia not observed. For illustrations of morphological characters, refer to Tibell et al. (2020).
Culture characteristics: Ascospores germinated on MEA within 24 h. Colonies on MEA, reaching 22–24 × 26–28 mm diam. in 25 days at 25 °C. Mycelium superficial, white to grayish white with age, hairy, effuse with wavy edge, dense, circular, raised, undulate to filiform with age; reverse dark yellowish brown.
Material examined: SWEDEN, Gotland, Kappelshamnsviken, on dead Phragmites culm (Poaceae), 7 March 2019, E.B.G. Jones, GJ654 (MFLU 20–0551), living culture MFLUCC 20–0224.
Notes: Keisslerialla linearis (MFLUCC 20–0224) groups with two strains of K. linearis (IFRD2008, MFLUCC 19–0410) with strong bootstrap support (100% ML, 100% MP, 1.00 BYPP; Fig. 1). The new strain is an asexual morph of K. linearis observed from Phragmites sp. in Sweden (Tibell et al. 2020). In the phylogenetic analysis, K. linearis clustered with other Keissleriella species. A comparison of the LSU and SSU sequence data of K. linearis (IFRD2008) and the new strain (MFLUCC 20–0224) revealed no nucleotide differences.
New combinations
Halobyssothecium cangshanense (Z.L. Luo, X.J. Su & K.D. Hyde) M.S. Calabon, K.D. Hyde & E.B.G. Jones, comb. nov.
Index Fungorum number: IF558092; Facesoffungi number: FoF 09434
Basionym: Lentithecium cangshanense Z.L. Luo, X.J. Su & K.D. Hyde, Phytotaxa 267 (1): 65 (2016)
Sexual morph: Descriptions and illustrations refer to Su et al. (2016). Asexual morph: Undetermined
Distribution: CHINA, Yunnan Province, saprobic on decaying wood submerged in a stream.
Notes: Holotype HKAS 84021. LSU and SSU sequence data are available.
Halobyssothecium carbonneanum (J. Fourn., Raja & Oberlies) M.S. Calabon, K.D. Hyde & E.B.G. Jones, comb. nov.
Index Fungorum number: IF558093; Facesoffungi number: FoF 09435
Basionym: Lentithecium carbonneanum J. Fourn., Raja & Oberlies, Persoonia 40: 295 (2018)
Sexual morph: Descriptions and illustrations refer to Crous et al. (2018). Asexual morph: Undetermined
Distribution: FRANCE, Haute-Garonne, Carbonne, SW of route du Lançon, artificial lake in a gravel pit, on submerged decorticated branch of Populus.
Notes: Holotype ILLS 81639. ITS, LSU and RPB2 sequence data are available.
Halobyssothecium kunmingense (W. Dong, H. Zhang & K.D. Hyde) M.S. Calabon, Boonmee, K.D. Hyde & E.B.G. Jones, comb. nov.
Index Fungorum number: IF556948; Facesoffungi number: FoF 09436
Basionym: Lentithecium kunmingense W. Dong, H. Zhang & K.D. Hyde
Sexual morph: Undetermined. Asexual morph: Descriptions and illustrations refer to Dong et al. (2020)
Distribution: CHINA, Yunnan Province, Kunming University of Science and Technology, on submerged wood in a stream (Dong et al. 2020).
Notes: HKAS 102150. LSU, SSU, ITS, TEF1-α sequence data are available.
Halobyssothecium unicellulare (Abdel-Aziz) M.S. Calabon, K.D. Hyde & E.B.G. Jones, comb. nov.
Index Fungorum number: IF558094; Facesoffungi number: FoF 09437
Basionym: Lentithecium unicellulare Abdel-Aziz, Fungal Diversity 80: 53 (2016)
Sexual morph: Undetermined. Asexual morph: Descriptions and illustrations refer to Hyde et al. (2016)
Distribution: EGYPT, Sohag City, on decayed wood submerged in the River Nile (Hyde et al. 2016).
Notes: Holotype CBS H-22674. LSU and SSU sequence data are available.
Halobyssothecium voraginesporum (Abdel-Wahab, Bahkali & E.B.G. Jones) M.S. Calabon, K.D. Hyde & E.B.G. Jones, comb. nov.
Index Fungorum number: IF558095; Facesoffungi number: FoF 09438
Basionym: Lentithecium voraginesporum Abdel-Wahab, Bahkali & E.B.G. Jones, Fungal Diversity 80: 53 (2016)
Sexual morph: Descriptions and illustrations refer to Hyde et al. (2016). Asexual morph: Undetermined
Distribution: SAUDI ARABIA, Arabian Gulf, Tarut mangroves, on submerged, decayed Phragmites australis (Poaceae), stem inside the mangrove stand (Hyde et al. 2016)
Notes: Holotype CBS H-22560. LSU and SSU sequence data are available.
Notes for Lentitheciaceae
Lentithecium aquaticum Ying Zhang, J. Fourn. & K.D. Hyde, Fungal Diversity 38: 234 (2009)
Phylogenetic analysis shows that Lentithecium aquaticum does not cluster within the Lentithecium clade but forms a weakly supported subclade basal to Darksidea species. Further collections are required to establish the taxonomic position of L. aquaticum.
Pseudomurilentithecium camporesii Mapook & K.D. Hyde, Fungal Diversity 100: 69 (2020)
In the phylogenetic analysis (Fig. 1), Psedomurilentithecium camporesii does not cluster within Lentitheciaceae but forms a weakly supported clade basal to Latoruaceae, Longipedicellataceae, and Trematosphaeriaceae. Broader taxon sampling, including other families in Pleosporales, is necessary to confirm its placement.
Keissleriella caudata (E. Müll.) Corbaz, Phytopathologische Zeitschrift 28 (4): 411 (1957)
Preliminary phylogenetic analysis shows that Keissleriella caudata does not group with other Keissleriella species, but clusters instead with Corynespora species. Only ITS sequence data of K. caudata is available in GenBank with an accession number MH857034. BLAST analysis did not show any Keissleriella species in the first 100 closely related sequence data. A fresh collection of specimens and additional DNA sequence data are required to confirm its placement within Pleosporales.
Discussion
Since Lentithecium was established for L. fluviatile (≡ Massarina fluviatilis), ten additional species have been introduced from lotic and lentic freshwater (Zhang et al. 2009b; Tanaka et al. 2015; Hyde et al. 2016; Su et al. 2016; Crous et al. 2018), as well as marine (Suetrong et al. 2009; Zhang et al. 2009b; Hyde et al. 2016) habitats and from different hosts. Lentithecium arundinaceum (≡ Massarina arundinacea), whose phylogenetic position was unclear for a long time and has been assigned to various genera (i.e., Ampullina, Heptameria, Leptosphaeria, Lophiostoma, Massarina, Metasphaeria, Peripherostoma, Phaeosphaeria, Pleospora, Rhopographus, Sphaeria, Sphaeropsis), was transferred by Tanaka et al. (2015) to Setoseptoria. Setoseptoria arundinacea clustered with other Setoseptoria species in the phylogenetic analysis (Fig. 1).
Multi-locus phylogenetic analysis shows that the three Lentithecium species, L. aquaticum, L. lineare and L. rarum (Kohlm., Volkm.-Kohlm. & O.E. Erikss.) Suetrong, Sakay., E.B.G. Jones, Kohlm. & Volkm.-Kohlm. do not group with other Lentithecium species, which was also reported by Tanaka et al. (2015), Devadatha et al. (2020), Dong et al. (2020), and Wijayawardene et al. (2020). Lentithecium aquaticum, a species introduced by Zhang et al. (2009b) based on LSU, SSU and RPB2 sequence data, forms a weakly supported clade basal to Darksidea and Lentithecium, which confirms the observations of Tanaka et al. (2015) (Fig. 1). Dayarathne et al. (2018) and Devadatha et al. (2020) showed that Lentithecium aquaticum clustered within Setoseptoria and the asexual morph Stagonospora macropycnidia, while Crous et al. (2018) confirmed that it does not group in Lentithecium.
Keissleriella rara was transferred to Lentithecium by Suetrong et al. (2009) together with K. cladophila and Massarina phragmiticola. The present phylogenetic analysis shows that Lentithecium rarum clustered in Keissleriella as sister taxon to K. trichophoricola Crous & Quaedvl. (Fig. 1). The same placement was observed also by Singtripop et al. (2015). Keissleriella linearis was transferred by Zhang et al. (2009b) to Lentithecium based on LSU and SSU sequence data. Keissleriella linearis, in common with other Keissleriella species, has short brown setae around the apex of the ascomatal ostiole, but Zhang et al. (2009b) opined that the presence of setae has little phylogenetic significance. In their phylogenetic analysis, other species and strains of Keissleriella were not included. Singtripop et al. (2015) reexamined the type specimen of L. lineare and transferred it to Keissleriella based on morphology and LSU sequence data, and this is in agreement with recent studies by Tanaka et al. (2015), Hyde et al. (2016) and the present study. However, Dayarathne et al. (2018) and Devadatha et al. (2020) placed L. lineare in the Lentithecium clade. The recent discovery of the asexual morph of L. lineare by Tibell et al. (2020) and the phylogenetic analysis based on the four-locus sequence dataset in the present study supports its taxonomic placement in Keissleriella.
The continuous discovery of novel fungal species has significantly contributed to the revision of fungal taxa (Arzanlou et al. 2007; Boonmee et al. 2011; Tanaka et al. 2015; Hashimoto et al. 2017; Hyde et al. 2018, 2020a,b,c). Phylogenetic analysis of the newly discovered Halobyssothecium species, including all the members of Lentitheciaceae, with molecular data supports the transfer of Lentithecium cangshanense, L. carbonneanum, L. kunmingense, L. unicellulare, and L. voraginesporum to Halobyssothecium. In the present placement, members of Halobyssothecium have brown and versicolored ascospores without sheath and hyaline conidia, while Lentithecium species possess hyaline ascospores with mucilaginous sheaths.
Key to Halobyssothecium species
-
1 Asexual morph...............................................................2
-
1* Sexual morph...............................................................5
-
2 Conidia, globose to obovate...........................................3
-
2* Conidia, ellipsoidal to cylindrical............H. phragmitis
-
3 Conidiomata > 350 μm long...................H. bambusicola
-
3* Conidiomata < 350 μm long........................................4
-
4 Conidiomata 210–250 × 320–350 μm...H. kunmingense
-
4* Conidiomata 115–235 × 140–235 μm...H. unicellulare
-
5 Ascospores, brown.........................................................6
-
5* Ascospores, versicolored.............................................8
-
6 Asci > 100 μm long.............................H. carbonneanum
-
6* Asci < 100 μm long......................................................7
-
7 Asci 38–50 × 8–10 μm......................H. voraginesporum
-
7* Asci 65–78 × 11–13 μm.....................H. cangshanense
-
8 Asci > 200 μm high........................................................9
-
8* Asci < 200 μm high...................................H. versicolor
-
9 Asci 180–214 × 12–16 μm.............................H. obiones
-
9* Asci 120–235 × 10–25 μm.........................H. estuariae
Data availability
All data generated or analyzed in this study are included in this article. All alignments and trees from this study are available from TreeBASE (accession number 27520) and all sequences generated here are available from GenBank with accession numbers: MT232434–MT232437, MN833419 (ITS); MT068485–MT068489 (LSU); MT068491–MT068494, MW346047 (SSU); MT477864–MT477868 (TEF1-α).
References
Apinis AE, Chesters CGC (1964) Ascomycetes of some salt marshes and sand dunes. Trans Br Mycol Soc 47:419–435. https://doi.org/10.1016/s0007-1536(64)80014-0
Arzanlou M, Groenewald JZ, Gams W et al (2007) Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud Mycol 58:57–93. https://doi.org/10.3114/sim.2007.58.03
Barr ME (1987) Prodromus to Class Loculoascomycetes. Amherst. University of Massachusetts, Massachusetts
Boonmee S, Zhang Y, Chomnunti P et al (2011) Revision of lignicolous Tubeufiaceae based on morphological reexamination and phylogenetic analysis. Fungal Divers 51:63–102. https://doi.org/10.1007/s13225-011-0147-4
Bruen TC, Philippe H, Bryant D (2006) A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681. https://doi.org/10.1534/genetics.105.048975
Cai L, Ji KF, Hyde KD (2006) Variation between freshwater and terrestrial fungal communities on decaying bamboo culms. Antonie van Leeuwenhoek. Int J Gen Mol Microbiol 89:293–301. https://doi.org/10.1007/s10482-005-9030-1
Cai L, Lumyong P, Zhang K, Hyde KD (2002a) New species of Annulatascus and Saccardoella from the Philippines. Mycotaxon 84:255–263
Cai L, Zhang K, Hyde KD (2005) Ascoyunnania aquatica gen. et sp. nov., a freshwater fungus collected from China and its microcylic conidiation. Fungal Divers 18:1–8. https://doi.org/10.2307/1468376
Cai L, Zhang K, McKenzie EHC et al (2002b) Acrodictys liputii sp. nov. and Digitodesmium bambusicola sp. nov. from bamboo submerged in the Liput River in the Philippines. Nova Hedwigia 75:525–532. https://doi.org/10.1127/0029-5035/2002/0075-0525
Cai L, Zhang K, McKenzie EHC, Hyde KD (2003) Freshwater fungi from bamboo and wood submerged in the Liput River in the Philippines. Fungal Divers 13:1–12.
Cai L, Zhang K, McKenzie EHC, Hyde KD (2004) Linocarpon bambusicola sp. nov. and Dictyochaeta curvispora sp. nov. from bamboo submerged in freshwater. Nova Hedwigia 78:439–445. https://doi.org/10.1127/0029-5035/2004/0078-0439
Calado M d L, Carvalho L, Pang KL, Barata M (2015) Diversity and ecological characterization of sporulating higher filamentous marine fungi associated with Spartina maritima (Curtis) Fernald in two Portuguese salt marshes. Microb Ecol 70:612–633. https://doi.org/10.1007/s00248-015-0600-0
Cavaliere AR (1968) Marine fungi of Iceland: A preliminary account of Ascomycetes. Mycologia 60:475–479. https://doi.org/10.2307/3757416
Cooke MC (1889) Synopsis Pyrenomycetum. Grevillea 18:28–33
Crouan PL, Crouan HM (1867) Florule du Finistère: Contenant les descriptions de 360 espèces nouvelles de sporagames, de nombreuses observations et une synonymie des plantes cellulaires et vasculaires qui croissent spontanément dans ce département; accompagnées de trente-deux planches où est représentée l’organographie, faite sur l’état vif. des fruits et des tissus de 198 genres d’algues avec la plante grandeur naturelle ou réduite plus une planche supplémentiare ou sont figurés 24 champignons nouveaux. F. Klincksieck, pp 22
Crous PW, Wingfield MJ, Burgess TI et al (2018) Fungal planet description sheets: 716–784. Persoonia Mol Phylogeny Evol Fungi 40:240–393. https://doi.org/10.3767/persoonia.2018.40.10
Crous PW, Wingfield MJ, Guarro J et al (2015) Fungal Planet description sheets: 320–370. Persoonia Mol Phylogeny Evol Fungi 34:167–266. https://doi.org/10.3767/003158515X688433
Cunnell GJ (1961) Stagonospora macropycnidia sp. nov. Trans Br Mycol Soc 44:87–90. https://doi.org/10.1016/s0007-1536(61)80010-7
Darriba D, Taboada GL, Doallo R, Posada D (2012) JModelTest 2: More models, new heuristics and parallel computing. Nat Methods 9:772
Dayarathne MC, Wanasinghe DN, Jones EBG et al (2018) A novel marine genus, Halobyssothecium (Lentitheciaceae) and epitypification of Halobyssothecium obiones comb. nov. Mycol Prog 17:1161–1171. https://doi.org/10.1007/s11557-018-1432-3
de Gruyter J, Aveskamp MM, Woudenberg JHC et al (2009) Molecular phylogeny of Phoma and allied anamorph genera: Towards a reclassification of the Phoma complex. Mycol Res 113:508–519. https://doi.org/10.1016/j.mycres.2009.01.002
Dennis RWG (1964) The Fungi of the Isle of Rhum. Kew Bull 19:77–127. https://doi.org/10.2307/4108295
Devadatha B, Calabon MS, Abeywickrama PD et al (2020) Molecular data reveals a new holomorphic marine fungus, Halobyssothecium estuariae, and the asexual morph of Keissleriella phragmiticola. Mycology 11:167–183. https://doi.org/10.1080/21501203.2019.1700025
Dong W, Wang B, Hyde KD et al (2020) Freshwater Dothideomycetes. Fungal Divers 105:319–575. https://doi.org/10.1007/s13225-020-00463-5
Glez-Peña D, Gómez-Blanco D, Reboiro-Jato M et al (2010) ALTER: Program-oriented conversion of DNA and protein alignments. Nucleic Acids Res 38:W14–W18. https://doi.org/10.1093/nar/gkq321
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41(1999):95–98. https://doi.org/10.14601/Phytopathol_Mediterr-14998u1.29
Hashimoto A, Matsumura M, Hirayama K, Tanaka K (2017) Revision of Lophiotremataceae (Pleosporales, Dothideomycetes): Aquasubmersaceae, Cryptocoryneaceae, and Hermatomycetaceae fam. nov. Persoonia Mol Phylogeny Evol Fungi 39:51–73. https://doi.org/10.3767/persoonia.2017.39.03
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42:182–192. https://doi.org/10.1093/sysbio/42.2.182
Hirayama K, Tanaka K, Raja HA et al (2010) A molecular phylogenetic assessment of Massarina ingoldiana sensu lato. Mycologia 102:729–746. https://doi.org/10.3852/09-230
Ho WH, Hyde KD, Hodgkiss IJ (2004) Cataractispora receptaculorum, a new freshwater ascomycete from Hong Kong. Mycologia 96:411–417. https://doi.org/10.1080/15572536.2005.11832986
Höhnel F (1919) Fragmente zur Mykologie. XXIII Mitteilung, Nr. 1154 bis 1188. Sitzungsberichte der Kaiserlichen Akademie der Wissenschaften Math.-naturw. Klasse Abt I 128:535–625
Hongsanan S, Hyde KD, Phookamsak R et al (2020) Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 11:1553–2107. https://doi.org/10.5943/mycosphere/11/1/13
Huson DH (1998) SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics 14:68–73. https://doi.org/10.1093/bioinformatics/14.1.68
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Hyde KD, Chethana KWT, Jayawardena RS et al (2020a) The rise of mycology in Asia. Sci Asia 46S:1–11. https://doi.org/10.2306/scienceasia1513-1874.2020.S001
Hyde KD, Dong Y, Phookamsak R et al (2020b) Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers 100:5–277. https://doi.org/10.1007/s13225-020-00439-5
Hyde KD, Hongsanan S, Jeewon R et al (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 80:1–270. https://doi.org/10.1007/s13225-016-0373-x
Hyde KD, Jeewon R, Chen YJ et al (2020c) The numbers of fungi: is the descriptive curve flattening? Fungal Divers 103:219–271. https://doi.org/10.1007/s13225-020-00458-2
Hyde KD, Jones EBG, Liu JK et al (2013) Families of Dothideomycetes. Fungal Divers 63:1–313. https://doi.org/10.1007/s13225-013-0263-4
Hyde KD, Mouzouras R (1988) Passeriniella savoryellopsis sp. nov., a new ascomycete from intertidal mangrove wood. Trans Br Mycol Soc 91:179–185. https://doi.org/10.1016/s0007-1536(88)80024-x
Hyde KD, Norphanphoun C, Chen J et al (2018) Thailand’s amazing diversity: up to 96% of fungi in northern Thailand may be novel. Fungal Divers 93:215–239. https://doi.org/10.1007/s13225-018-0415-7
Jayasiri SC, Hyde KD, Ariyawansa HA et al (2015) The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers 74:3–18. https://doi.org/10.1007/s13225-015-0351-8
Jeewon R, Hyde KD (2016) Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 7:1669–1677. https://doi.org/10.5943/mycosphere/7/11/4
Jones EBG, Pang KL, Abdel-Wahab MA et al (2019) An online resource for marine fungi. Fungal Divers 96:347–433. https://doi.org/10.1007/s13225-019-00426-5
Jones EBG (1962) Marine fungi. Trans Br Mycol Soc 45:93–114
Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. https://doi.org/10.1093/bib/bbx108
Khashnobish A, Shearer CA (1996) Reexamination of some Leptosphaeria and Phaeosphaeria species, Passeriniella obiones and Melanomma radicans. Mycol Res 100:1341–1354. https://doi.org/10.1016/S0953-7562(96)80062-1
Knapp DG, Kovács GM, Zajta E et al (2015) Dark septate endophytic pleosporalean genera from semiarid areas. Persoonia Mol Phylogeny Evol Fungi 35:87–100. https://doi.org/10.3767/003158515X687669
Kohlmeyer J, Volkmann-Kohlmeyer B, Eriksson OE (1996) Fungi on Juncus roemerianus. New marine and terrestrial ascomycetes. Mycol Res 100:393–404. https://doi.org/10.1016/S0953-7562(96)80134-1
Kohlmeyer J, Kohlmeyer E (1979) Marine mycology: The higher fungi. Academic Press, New York
Li GJ, Hyde KD, Zhao RL et al (2016) Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 78:1–237. https://doi.org/10.1007/s13225-016-0366-9
Liu JK, Hyde KD, Jeewon R et al (2017) Ranking higher taxa using divergence times: a case study in Dothideomycetes. Fungal Divers 84:75–99. https://doi.org/10.1007/s13225-017-0385-1
Lumbsch HT, Huhndorf SM (2010) Myconet Volume 14. Part One. Outline of Ascomycota—2009. Fieldiana Life Earth Sci 1:1–922. https://doi.org/10.3158/1557.1
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway computing environments workshop. GCE 2010. https://doi.org/10.1109/GCE.2010.5676129
Nylander JAA (2004) MrModeltest 2.0. Program distributed by the author. Evolutionary Biology Centre. Uppsala University, Uppsala
Phookamsak R, Manamgoda DS, Li WJ et al (2015) Poaceascoma helicoides gen. et sp. nov., a new genus with scolecospores in Lentitheciaceae. Cryptogam Mycol 36:225–236. https://doi.org/10.7872/crym/v36.iss2.2015.225
Quaedvlieg W, Verkley GJM, Shin HD et al (2013) Sizing up Septoria. Stud Mycol 75:307–390. https://doi.org/10.3114/sim0017
Quaedvlieg W, Binder M, Groenewald JZ et al (2014) Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia Mol Phylogeny Evol Fungi 33:1–40. https://doi.org/10.3767/003158514X681981
Rehner S (2001) Primers for Elongation Factor 1-alpha (EF1-alpha). Insect Biocontrol Laboratory: USDA, ARS, PSI rehner@ba.ars.usda.gov.
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Saccardo PA (1882) Fungi boreali-americani. Michelia 2:564–582
Saccardo PA (1883) Sylloge Pyrenomycetum, Vol. II. Sylloge Fungorum 2:1–813
Schoch CL, Crous PW, Groenewald JZ et al (2009) A class-wide phylogenetic assessment of Dothideomycetes. Stud Mycol 64:1–15-S10. https://doi.org/10.3114/sim.2009.64.01
Singtripop C, Camporesi E, Ariyawansa HA et al (2015) Keissleriella dactylidis, sp. nov., from Dactylis glomerata and its phylogenetic placement. ScienceAsia 41:295–304. https://doi.org/10.2306/scienceasia1513-1874.2015.41.295
Stamatakis A (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. https://doi.org/10.1093/bioinformatics/btl446
Stamatakis A (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57:758–771. https://doi.org/10.1080/10635150802429642
Su HY, Luo ZL, Liu XY et al (2016) Lentithecium cangshanense sp. nov. (Lentitheciaceae) from freshwater habitats in Yunnan Province, China. Phytotaxa 267:61–69. https://doi.org/10.11646/phytotaxa.267.1.6
Suetrong S, Schoch CL, Spatafora JW et al (2009) Molecular systematics of the marine Dothideomycetes. Stud Mycol 64:155–173.S6. https://doi.org/10.3114/sim.2009.64.09
Swofford DL (2002) Phylogenetic analysis using parsimony. Options 42:294–307. https://doi.org/10.1007/BF02198856
Tanaka K, Harada Y (2005) Bambusicolous fungi in Japan (6): Katumotoa, a new genus of phaeosphaeriaceous ascomycetes. Mycoscience 46:313–318. https://doi.org/10.1007/s10267-005-0251-y
Tanaka K, Hatakeyama S, Harada Y (2005) Three new freshwater ascomycetes from rivers in Akkeshi, Hokkaido, northern Japan. Mycoscience 46:287–293. https://doi.org/10.1007/s10267-005-0248-6
Tanaka K, Hirayama K, Yonezawa H et al (2015) Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud Mycol 82:75–136. https://doi.org/10.1016/j.simyco.2015.10.002
Tibell S, Tibell L, Pang KL et al (2020) Marine fungi of the Baltic Sea. Mycology 11:195–213. https://doi.org/10.1080/21501203.2020.1729886
Tibpromma S, Hyde KD, Jeewon R et al (2017) Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 83:1–261. https://doi.org/10.1007/s13225-017-0378-0
Van Ryckegem G, Aptroot A (2001) A new Massarina and a new Wettsteinina (Ascomycota) from freshwater and tidal reeds. Nova Hedwigia 73:161–166. https://doi.org/10.1127/nova.hedwigia/71/2001/161
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
Wanasinghe DN, Jones EBG, Camporesi E et al (2014) An exciting novel member of Lentitheciaceae in Italy from Clematis vitalba. Cryptogam Mycol 35:323–337. https://doi.org/10.7872/crym.v35.iss4.2014.323
Webber EE (1970) Marine ascomycetes from New England. Bull Torrey Bot Club 97:119–120. https://doi.org/10.2307/2483402
White TJ, Bruns T, Lee S, Taylo JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
Wijayawardene NN, Hyde KD, Bhat DJ et al (2015) Additions to brown spored coelomycetous taxa in Massarinae, Pleosporales: introducing Phragmocamarosporium gen. nov. and Suttonomyces gen. nov. Cryptogam Mycol 36:213–224. https://doi.org/10.7872/crym/v36.iss2.2015.213
Wijayawardene NN, Hyde KD, Al-Ani LKT et al (2020) Outline of Fungi and fungus-like taxa. Mycosphere 11:1–367. https://doi.org/10.5943/mycosphere/11/1/8
Zhang H, Dong W, Hyde KD et al (2017) Towards a natural classification of Annulatascaceae-like taxa: introducing Atractosporales ord. nov. and six new families. Fungal Divers 85:75–110. https://doi.org/10.1007/s13225-017-0387-z
Zhang Y, Crous PW, Schoch CL, Hyde KD (2012) Pleosporales. Fungal Divers 53:1–221. https://doi.org/10.1007/s13225-011-0117-x
Zhang Y, Schoch CL, Fournier J et al (2009a) Multi-locus phylogeny of Pleosporales: A taxonomic, ecological and evolutionary re-evaluation. Stud Mycol 64:85–102.S5. https://doi.org/10.3114/sim.2009.64.04
Zhang Y, Wang HK, Fournier J et al (2009b) Towards a phylogenetic clarification of Lophiostoma/Massarina and morphologically similar genera in the Pleosporales. Fungal Divers 38:225–251
Acknowledgments
MS Calabon is grateful to Mushroom Research Foundation and Department of Science and Technology – Science Education Institute (Philippines). The authors are grateful to Dr. Eleni Gentekaki, Dr. Bandarupalli Devadatha, Dr. Mingkwan Doilom, Chun-Fang Liao, and Er-Fu Yang for their comments and suggestions to improve the manuscript, and for helping with the molecular analysis of the samples. W. Dong, H. Zhang, and K.D. Hyde are thanked for the release of data on Lentithecium kunmingense W. Dong, H. Zhang & K.D. Hyde prior to publication of their paper.
Funding
Mark Calabon is grateful to the 5th batch of Postdoctoral Orientation Training Personnel in Yunnan Province (grant no.: Y934283261) and the 64th batch of China Postdoctoral Science Foundation (grant no.: Y913082271). E. B Gareth Jones is supported under the Distinguished Scientist Fellowship Program (DSFP), King Saud University, Kingdom of Saudi Arabia. The Swedish Species Initiative (“ArtDatabanken”) is thanked for support within the project “Marine fungi in Sweden” (SLU.dha.2017.4.3-73). Kevin D. Hyde thanks the Thailand Research Fund grant entitled “Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion” (Grant No. RDG6130001). Rungtiwa Phookamsak is supported by CAS President’s International Fellowship Initiative (PIFI) for Young Staff 2019–2021 (grant number 2019FY0003), The Yunnan Provincial Department of Human Resources and Social Security (Grant No. Y836181261), and The National Science Foundation of China (NSFC) project code 31850410489.
Author information
Authors and Affiliations
Contributions
Conceptualization: Mark S. Calabon, E.B. Gareth Jones, and Kevin D. Hyde. Methodology: Mark S. Calabon, E.B. Gareth Jones, and Kevin D. Hyde. Formal analysis and investigation: Mark S. Calabon, E.B. Gareth Jones, Kevin D. Hyde, Ka-Lai Pang, and Sanja Tibell. Resources: E.B. Gareth Jones, Kevin D. Hyde, and Rungtiwa Phookamsak. Writing—original draft preparation: Mark S. Calabon. Writing—review and editing: E.B. Gareth Jones, Kevin D. Hyde, Saranyaphat Boonmee, Sanja Tibell, Leif Tibell, Ka-Lai Pang, and Rungtiwa Phookamsak. Supervision: E.B. Gareth Jones and Kevin D. Hyde. Funding acquisition: E.B. Gareth Jones, Kevin D. Hyde, Saranyaphat Boonmee, and Rungtiwa Phookamsak. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Roland Kirschner
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Calabon, M.S., Jones, E.G., Hyde, K.D. et al. Phylogenetic assessment and taxonomic revision of Halobyssothecium and Lentithecium (Lentitheciaceae, Pleosporales). Mycol Progress 20, 701–720 (2021). https://doi.org/10.1007/s11557-021-01692-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-021-01692-x