Abstract
During 2017, the Westerdijk Fungal Biodiversity Institute (WI) and the Utrecht University Museum launched a Citizen Science project. Dutch school children collected soil samples from gardens at different localities in the Netherlands, and submitted them to the WI where they were analysed in order to find new fungal species. Around 3000 fungal isolates, including filamentous fungi and yeasts, were cultured, preserved and submitted for DNA sequencing. Through analysis of the ITS and LSU sequences from the obtained isolates, several plectosphaerellaceous fungi were identified for further study. Based on morphological characters and the combined analysis of the ITS and TEF1-α sequences, some isolates were found to represent new species in the genera Phialoparvum, i.e. Ph. maaspleinense and Ph. rietveltiae, and Plectosphaerella, i.e. Pl. hanneae and Pl. verschoorii, which are described and illustrated here.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil is one of the main reservoirs of fungal species and commonly ranks as the most abundant source regarding fungal biomass and physiological activity. Fungal diversity is affected by the variety of microscopic habitats and microenvironments present in soils (Anderson and Domsch 1978; Bills et al. 2004). More than half of the soil mycobiota is composed of basidiomycetes, although they are overlooked by most isolation techniques (Kirk et al. 2001). The remaining fungal biomass is comprised by Ascomycota, Chytridiomycota, Mucorales and Oomycetes (Kirk et al. 2001; Bills et al. 2004; Crous et al. 2019). This observed species richness has been supported by the discovery of previously unrecognised phylogenetic fungal lineages in soil-inhabiting fungi (Tedersoo et al. 2017).
Among Ascomycota, the family Plectosphaerellaceae (Glomerellales, Sordariomycetes) harbours important plant pathogens such as Verticillium dahliae, V. alboatrum and Plectosphaerella cucumerina, but also several saprobic genera usually found in soil, i.e. Chordomyces, Gibellulopsis and Sodiomyces (Domsch et al. 2007; Zare et al. 2007; Carlucci et al. 2012; Grum-Grzhimaylo et al. 2013, 2016). Members of this family are mainly known from their asexual morphs, which are morphologically characterised by simple or verticillate conidiophores with mono- or polyphialidic conidiogenous cells, and 1- or 2-celled elongate conidia arranged in slimy heads or chains, and rarely produced sporodochia or synnemata. The sexual morph is mostly observed in culture, showing perithecial or cleistothecial ascomata, superficial, brown to dark brown, with clavate or saccate asci and hyaline to pale brown ascospores (Giraldo and Crous 2019).
The interest in soil fungi has recently increased due to the high demand for new compounds with application in the biotechnological or pharmaceutical industries (Hujslová et al. 2010), and also because this substratum contains a pool of interesting and undescribed species, especially when samples from diverse habitats are studied (Blackwell 2011; Tedersoo et al. 2014, 2017). Therefore, during 2017, children from different locations in the Netherlands collected soil samples which were analysed in order to find new fungal species. This was done during the course of a Citizen Science project, named ‘Wereldfaam, een schimmel met je eigen naam’ (World fame - a fungus with your name) coordinated by the Westerdijk Fungal Biodiversity Institute and the Utrecht University Museum. Among the examined samples, several plectosphaerellaceous fungi were isolated, some of which have been recently described as new species in the genera Gibellulopsis and Lectera (Crous et al. 2018), while others represent new species in the genera Phialoparvum and Plectosphaerella, which are described and illustrated here.
Materials and methods
Sampling and isolation
Protocols for the collection and processing of soil samples are described in Groenewald et al. (2018). After 1 week of incubation on MEA supplemented with penicillin-G and streptomycin, individual colonies were transferred to MEA plates without antibiotics and incubated between 22 and 24 °C for 7–14 days, in order to obtain axenic cultures. Subsequently, each isolate was submitted for DNA extraction and sequencing. All isolates are maintained in the Johanna Westerdijk (JW) collection housed at the Westerdijk Fungal Biodiversity Institute in Utrecht, The Netherlands. Isolates from new and interesting species are also deposited at the CBS Culture Collection at the WI.
DNA extraction, amplification and sequencing
Total genomic DNA was extracted from colonies growing on MEA using the Wizard® Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA), following the manufacturer’s protocol. The internal transcribed spacer (ITS) regions and the 5′ end of the 28S nrDNA gene (LSU) were amplified for all isolates with the primer combination ITS5/LR5, and sequenced with the primer pairs ITS5/ITS4 (White et al. 1990) and LR0R/LR5 (Vilgalys and Hester 1990; Vilgalys and Sun 1994), respectively. Additional loci were sequenced for some isolates, including fragments from the protein-coding genes translation elongation factor 1-alpha (TEF1-α) and RNA polymerase II second largest subunit (RPB2) with the primer sets EF-983F/EF-2218R (Rehner and Buckley 2005) and RPB2-5F2/RPB2-7cR (Liu et al. 1999), respectively. The consensus sequence of each locus was obtained by using the program SeqMan v. 12.1.0 (DNASTAR, Madison, WI, USA).
Species identification
Firstly, the LSU and ITS consensus sequences from each isolate were blasted against sequences available in NCBI (http://blast.ncbi.nlm.nih.gov) and CBS (http://www.westerdijkinstitute.nl/Collections) databases, in order to determine the generic placement of each isolate. Secondly, sequences from all isolates identified as members of the Plectosphaerellaceae were aligned with sequences from ex-type and reference strains of the species currently accepted in the genera (Table 1) to which each isolate belonged. Separate analyses based on ITS or ITS combined with TEF1-α were performed for those isolates that were not identified at level species by using the previous approach.
Phylogenetic analysis
Sequences were aligned through MAFFT v. 7 (Katoh et al. 2017), using the default parameters, and were manually corrected in MEGA v. 6.06 (Tamura et al. 2013). Phylogenetic inferences of the individual gene regions and the combined dataset were based on maximum likelihood (ML) and were performed on the CIPRES Science Gateway portal (Miller et al. 2012) using RAxML v. 8.2.10 (Stamatakis, 2014). The default parameters were used, and bootstrap support (BS) was carried out using the rapid bootstrapping algorithm with the automatic halt option. A BS ≥ 70% was considered as statistically significant. Each partition was assessed for incongruence before being concatenated by checking individual phylogenies for conflicts between clades with significant ML support (Mason-Gamer and Kellogg 1996; Wiens 1998). All novel DNA sequences generated in this study were deposited in GenBank and ENA databases (Table 1), while the alignments and the resulting trees were accessioned in TreeBASE (http://www.treebase.org).
Morphological characterisation of novel species
Morphological features from the new species were determined on oatmeal agar (OA), potato carrot agar (PCA), 2% potato dextrose agar (PDA) and 2% malt extract agar (MEA) (recipes in Crous et al. 2019). Cultures were incubated at ca. 24 °C in the dark for 4 weeks. Macroscopic characters and diameters were measured after 14 days of incubation, and the colony colour (surface and reverse) rated after Rayner (1970). Microscopic features were examined from slide cultures and preparations mounted in clear lactic acid from colonies sporulating on the media previously mentioned. Observations were performed with a Zeiss V20 Discovery (Zeiss, Oberkochen, Germany) stereo microscope and with a Zeiss Axio Imager 2 light microscope using differential interference contrast (DIC) optics. Photomicrographs and measurements were taken with a Nikon DS-Ri2 digital camera (Nikon, Tokyo, Japan) using the NIS-elements imaging software v. 4.3. The length and width of at least 30 randomly selected structures were measured, and the extreme values calculated. Morphological descriptions and taxonomic information for the new species were deposited in MycoBank (www.MycoBank.org; Crous et al. 2004).
Results
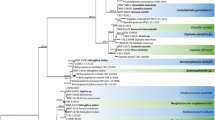
A total of 293 soil samples were analysed, and nearly 3000 fungal isolates were obtained. Among them, 176 were identified as members of Plectosphaerellaceae according to the BLAST search results using the LSU and/or ITS sequences. The combined ITS and LSU dataset comprises 197 sequences including the ones generated in the present study from soil isolates, together with reference sequences of Plectosphaerellaceae taxa download from GenBank, and the outgroup Monilochaetes infuscans CBS 379.77 and CBS 869.96 (Fig. 1). The RAxML tree showed BS values higher than 95% for all the generic clades, except for Gibellulopsis and Brunneochlamydosporum that were not fully supported (Fig. 1). The majority of the isolates were distributed among Plectosphaerella spp. (n = 61) and Gibellulopsis spp. (n = 60). They were followed by Chordomyces spp. (n = 22), Acrostalagmus luteoalbus (n = 15) and Brunneochlamydosporium spp. (n = 9). The remaining isolates were identified as Furcasterigmium furcatum (n = 4), Lectera nordwiniana (n = 3) and Phialoparvum spp. (n = 2) (Table 1). Since the species boundaries were not resolved with this analysis for Brunneochlamydosporium, Chordomyces, Gibellulopsis, Phialoparvum and Plectosphaerella, subsequent phylogenetic analyses were carried out (Figs. 2, 3, 4 and 5).
Maximum composite likelihood tree based on partial sequences from the ITS and LSU from all the soil isolates placed in Plectosphaerellaceae. Colour boxes indicate the generic clade. Bootstrap support values above 70% are shown at the genus nodes (internal values are not shown). TEx-type, ITEx-isotype, NTEx-neotype
The Brunneochlamydosporium dataset comprises ITS and TEF1-α sequences of 19 isolates including the outgroups (Fig. 2). This combined analysis showed B. nepalense (n = 7) as the most common species, followed by B. macroclavatum (n = 1). The ITS phylogeny represented in Fig. 3 shows the species distribution from all the isolates belonging to Chordomyces, Lectera and Phialoparvum. In the Chordomyces clade (95% BS), most of the isolates (n = 17) were grouped with the type of C. albus CBS 987.87 (76%), while the remaining isolates (n = 5) were identified as C. antarticus. The three isolates of Lectera were confirmed as L. nordwiniana, but the two Phialoparvum isolates were genetically different from the type of Ph. bifurcatum CBS 299.70B, representing two putative new species, introduced in the ‘Taxonomy’ section.
The phylogenetic analysis shown in Fig. 4 encompasses the ITS sequences from 106 isolates belonging to Gibellulopsis (including the ex-type strains from the different Gibellulopsis species), and Furcasterigmium and Musidium as outgroups (Fig. 4). According to this phylogeny, the majority of the soil isolates were identified as G. nigrescens (n = 50), followed by G. fusca (n = 5), G. serrae (formerly G. piscis) (n = 3) and G. simonii (n = 2).
A preliminary phylogenetic analysis based on the ITS region was carried out for Plectosphaerella (data not shown). Species were distributed in Pl. plurivora (n = 37), Pl. cucumerina (n = 9) and Pl. pauciseptata (n = 1) (Table 1). However, 14 isolates were distributed in two different lineages (with 1 and 13 isolates, respectively), phylogenetically distant from the other species. Subsequently, isolates of each clade were selected based in the similarity of their ITS regions for further combined analysis of ITS and TEF1-α sequences. This analysis included 55 soil isolates and 32 reference strains (Fig. 5), and confirmed the identity of the isolates placed in the Pl. plurivora (77% BS) and Pl. cucumerina clades (88% BS). Furthermore, this analysis revealed that 13 isolates were nested in a highly supported clade (99% BS), phylogenetically distant from the remaining species in the genus, representing a potentially new species. Isolate JW 181001 was placed in a single branch related (71% BS) but different from Pl. alismatis, Pl. delsorboi, Pl. melonis and Pl. sinensis. The two new species are described in the ‘Taxonomy’ section below.
Taxonomy
Based on the molecular results and the morphological observations, four new species are described and illustrated here, i.e. Phialoparvum maaspleinense, Ph. rietveltiae, Plectosphaerella hanneae and Pl. verschoorii.
Phialoparvum maaspleinense Hern.-Restr. & Giraldo López, sp. nov. MycoBank MB 831346 (Fig. 6a, b, e–j).
Phialoparvum spp. a, b, e–jPhialoparvum maaspleinense (ex-type CBS 145321 = JW 266001). a, b Colonies on PDA and MEA respectively, after 14 days at ca. 24 °C. e, f Monophialides and conidia. g Polyphialide and conidium. h, i Adelophialides and conidia. j Conidia. c, d, k–nPhialoparvum rietveltiae (ex-type CBS 145322 = JW 211005). c, d Colonies on PDA and MEA respectively, after 14 days at ca. 24 °C. k Monophialides and conidia. l Polyphialides and conidia. m Adelophialide. n Conidia. Scale bars: g–i, m = 5 μm. e, f, j–l, n = 10 μm
Etymology. Named after ‘Maaspleinschool’ from Utrecht, where the soil sample was collected by the students Ouail Zaim and Mohamed Bidari.
Mycelium consisting of branched, septate, smooth, hyaline and thick-walled hyphae, 1.5–2.5 μm wide. Conidiophores solitary, erect, arising directly from vegetative hyphae or ropes of hyphae, unbranched or poorly branched. Phialides lateral, terminal, subulate, hyaline, thick- and smooth-walled, 10–42 × 1.5–3 μm, with cylindrical collarette and conspicuous periclinal thickening at the conidiogenous locus; adelophialides commonly present, intercalary and terminal, up to 4 μm long; polyphialides with two conidiogenous loci are commonly present. Conidia arranged in slimy heads, cylindrical, sometimes with slightly truncate base, 1-celled, hyaline, thick- and smooth-walled, 3.5–5 × 2 μm. Sexual morph unknown.
Culture characteristics — After 14 days at ca. 24 °C on PDA reaching 45–48 mm diam., flat, membranous, aerial mycelium sparse, dirty white, reverse uncoloured. On OA reaching 40–42 mm diam., flat, pulverulent, rosy buff, margin effuse, reverse buff to honey. On MEA reaching 37–38 mm diam., radially folded, velvety at centre, pulverulent toward the periphery, dirty white, margin entire, reverse uncoloured.
Typus. The Netherlands, Utrecht province, Utrecht, Maaspleinschool, from garden soil, 2017, coll. O. Zaim & M. Bidari, isol. A. Giraldo (holotype CBS H-23910, cultures ex-type CBS 145321 = JW 266001).
Notes: The monotypic genus Phialoparvum was recently proposed based on Ph. bifurcatum, recovered from soil in Belgium (Giraldo and Crous 2019). Phialoparvum bifurcatum and the new species, Ph. maaspleinense can be morphologically distinguished based on phialide length (up to 15 μm in Ph. bifurcatum vs up to 42 μm in Ph. maaspleinense; Giraldo and Crous 2019) and the frequent production of intercalary and terminal adelophialides in the latter species (Fig. 6h, i).
Phialoparvum rietveltiae Hern.-Restr. & Giraldo López, sp. nov. MycoBank MB 831347 (Fig. 6c, d, k–n).
Etymology. Named after Emma Rietvelt, who collected the soil sample.
Mycelium consisting of branched, septate, hyaline and thick-walled hyphae, 1–2 μm wide. Conidiophores solitary, erect, arising directly from vegetative hyphae or ropes of hyphae, unbranched or poorly branched. Phialides lateral, terminal, subulate to ampulliform, hyaline, thick- and smooth-walled, 6–11(− 17) × 1–2.5 μm, with cylindrical collarette and conspicuous periclinal thickening at the conidiogenous locus, adelophialides sometimes present, up to 4 μm long; polyphialides with two conidiogenous loci are commonly present. Conidia arranged in slimy heads, cylindrical, with slightly pointed ends, 1-celled, hyaline, thick- and smooth-walled, 3.5–5 × 1.5–2 μm. Sexual morph unknown.
Culture characteristics — After 14 days at ca. 24 °C on PDA reaching 30–33 mm diam., flat, membranous, aerial mycelium sparse or absent, dirty white, margin entire, reverse uncoloured. On OA reaching 28–30 mm diam., flat, pulverulent to velvety, dirty white to buff, margin rhizoid, reverse uncoloured. On MEA reaching 24–29 mm diam., umbonate, radially folded, membranous to felty, buff, entire margin, reverse uncoloured.
Typus. The Netherlands, Utrecht province, Utrecht, from garden soil, 2017, coll. E. Rietvelt, isol. A. Giraldo (holotype CBS H-23911, cultures ex-type CBS 145322 = JW 211005).
Notes: Phialoparvum bifurcatum and Ph. rietveltiae are morphologically similar in phialide shape and length (8–15 μm in Ph. bifurcatum vs 6–17 μm in Ph. rietveltiae). However, the latter species has longer conidia (3.5–5 μm in Ph. rietveltiae vs 2.8–4.4 μm in Ph. bifurcatum) and does not produce melanin precipitations in culture (Giraldo and Crous 2019).
Plectosphaerella hanneae Giraldo López & Hern.-Restr., sp. nov. MycoBank MB 831348 (Fig. 7a–d).
Plectosphaerella spp. a–dPlectosphaerella hanneae (ex-type CBS 144925 = JW 181001). a Colony on PDA after 14 days at ca. 24 °C. b Monophialide and conidia. c Monophialides with a percurrent proliferation (arrow) and conidia. d Septate and aseptate conidia. e–iPlectosphaerella verschoorii (ex-type CBS 144924 = JW 13004). e Colony on PDA after 14 days at ca. 24 °C. f Monophialides and conidia. g Adelophialide and conidia. h, i Septate and aseptate conidia. Scale bars = 10 μm
Etymology. Named after Hanne de Levita, who collected the soil sample.
Mycelium consisting of branched, septate, smooth, hyaline and thin-walled hyphae, up to 2 μm wide. Conidiophores solitary, erect, unbranched, hyaline, smooth-walled, sometimes radiating out from sterile coils formed by the mycelium. Phialides lateral or terminal, subulate, hyaline, smooth- and thick-walled, occasionally borne on short cylindrical subtending cells, 16–32 × 2–4.5 μm, with conspicuous cylindrical collarette and periclinal thickening at the conidiogenous locus, sometimes with percurrent proliferation; adelophialides rarely present, up to 15 μm long. Conidia arranged in slimy heads, (0-)1-septate, cylindrical to ellipsoidal with pointed apex and slightly truncate base, inequilateral, with inner plane flat and outer plane convex, guttulate, hyaline, thin- and smooth-walled; septate conidia abundant, 9–12.5 × 2.5–3.5 μm; aseptate conidia scarce, 9–10.5 × 3–3.5 μm. Chlamydospores and sexual morph absent.
Culture characteristics — After 14 days at ca. 24 °C on PDA reaching 76–77 mm diam., flat, membranous, dirty white to pale luteous, aerial mycelium sparse or absent, with concentric rings, reverse uncoloured. On OA reaching 58–60 mm diam., flat, glabrous, pale luteous with ochreous border, aerial mycelium sparse at the periphery, reverse uncoloured.
Typus. The Netherlands, South Holland, Leiden, from garden soil, 2017, coll. H. de Levita, isol. A. Giraldo (holotype CBS H-23737, cultures ex-type CBS 144925 = JW 181001).
Notes: According to phylogenetic inference from the ITS and TEF1-α loci (Fig. 5), Pl. hanneae occupies a single branch in the clade containing Pl. alismatis, Pl. delsorboi, Pl. melonis and Pl. sinensis (71% BS). With the exception of Pl. delsorboi and Pl. hanneae, all the species in that clade produce hyaline chlamydospores in chains (Su et al. 2017). Plectosphaerella delsorboi differs in having longer phialides (30–50 μm; Antignani et al. 2008) than those of Pl. hanneae (16–32 μm).
Plectosphaerella verschoorii Giraldo López & Hern.-Restr., sp. nov. MycoBank MB 831349 (Fig. 7e–i).
Etymology. Named after Brent Verschoor, who collected the soil sample.
Mycelium consisting of branched, septate, smooth, hyaline and thin-walled hyphae, up to 2 μm wide. Conidiophores solitary, erect, unbranched or poorly branched, hyaline, smooth-walled. Phialides lateral or terminal, subcylindrical to subulate, hyaline, smooth- and thick-walled, 16–29 × 1.5–3 μm, with conspicuous cylindrical collarette and periclinal thickening at the conidiogenous locus; adelophialides, rarely present; ampulliform, up to 14 μm long. Conidia arranged in slimy head, (0-)1-septate, cylindrical to ellipsoidal with pointed apex and slightly truncate base, or broadly ellipsoidal, guttulate, hyaline, thin- and smooth-walled; septate conidia abundant, sometimes slightly constricted at the septum, 8–11.5 × 2–3 μm; aseptate conidia scarce, 3–8.5 × 2–3 μm. Chlamydospores and sexual morph absent.
Culture characteristics — After 14 days at ca. 24 °C on PDA reaching 76–77 mm diam., flat, membranous, flesh at centre, pale luteous at periphery, aerial mycelium sparse, with concentric rings, dirty white, reverse uncoloured. On OA reaching 70–75 mm diam., flat, glabrous, honey, reverse uncoloured.
Typus. The Netherlands, North Holland, Alkmaar, from garden soil, 2017, coll. B. Verschoor, isol. A. Giraldo (holotype CBS H-23738, cultures ex-type CBS 144924 = JW 13004).
Additional specimens examined: The Netherlands, Limburg, Nederweert, from garden soil, 2017, coll. M. van Meijl, isol. A. Giraldo, JW 232007; North Brabant, Liempde, from garden soil, 2017, coll. L. & V. Gijzen, isol. A. Giraldo, JW 150001; North Holland, Alkmaar, from garden soil, 2017, coll. Brent Verschoor, isol. A. Giraldo, JW 13006; Heemstede, from garden soil, 2017, coll. Alfons Vaessen, isol. A. Giraldo, JW 146007, JW 146009, JW 146016; Hilversum, from garden soil, 2017, coll. J.A.L. Keyes-Rens, isol. A. Giraldo, JW 208020; South Holland, Delft, from garden soil, 2017, E. & M. Bordes, isol. A. Giraldo, JW 62006; Utrecht, Amersfoort, from garden soil, 2017, coll. T. & K. Wesselink, isol. A. Giraldo, JW 191043; Maarssen, from garden soil, 2017, coll. Y. & F. van der Ouderaa, isol. A. Giraldo, JW 230011; Utrecht, from garden soil, 2017, coll. G. Bleijlevens, isol. A. Giraldo, JW 143005; from garden soil, 2017, coll. J.P. van Eesteren, isol. A. Giraldo, JW 170008.
Notes: Plectosphaerella verschoorii forms a well-supported clade (99% BS, Fig. 5), phylogenetically different from other Plectosphaerella species. This species was collected in five provinces from The Netherlands (i.e. Limburg, North Brabant, North and South Holland, and Utrecht).
Morphologically, Plectosphaerella verschoorii resembles Pl. citrullae, Pl. delsorboi, Pl. hanneae and Pl. plurivora in lacking polyphialides. However, it can be distinguished from Pl. plurivora by the scarce production of aseptate conidia; from Pl. delsorboi and Pl. citrullae by having shorter phialides (up to 29 μm in Pl. verschoorii, up to 50 μm in Pl. delsorboi, up to 60 μm in Pl. citrullae; Carlucci et al. 2012; Liu et al. 2013), and from Pl. hanneae by producing smaller aseptate conidia (3–8.5 × 2–3 μm in Pl. verschoorii vs. 9–10.5 × 3–3.5 μm in Pl. hanneae, this study).
Discussion
Although there are no specific studies exploring the diversity of the plectosphaerellaceous species from soil, a survey about fungi from soda soils in Asia (Armenia, Kazakhstan, Mongolia and Russia) and Africa (Kenia and Tanzania) revealed several isolates representing four genera of Plectosphaerellaceae (Grum-Grzhimaylo et al. 2016). That study showed Sodiomyces spp. and A. luteoalbus as obligate and facultative alkaliphilic fungi, respectively, while C. antarticus and V. zaregamsianum seemed to be alkalitolerant species. In our set of samples, we did not find Sodiomyces spp. nor V. zaregamsianum, probably due of the media used for primary isolation. However, in our study, A. luteoalbus and C. antarticus were represented by 13 and five isolates respectively (Figs. 1 and 3 and Table 1). Additionally, we obtained several isolates (n = 17) of C. albus, which was recently described from a lichen (Hypogymnia physodes) and human sputum (Giraldo et al. 2017), but also reported from soil in Belgium, France, Germany and the Netherlands (Giraldo and Crous 2019).
In addition, we have identified several isolates from genera recently added to Plectosphaerellaceae, i.e. Brunneochlamydosporium, Furcasterigmium, Lectera and Phialoparvum (Cannon et al. 2012; Giraldo and Crous 2019). Brunneochlamydosporium was proposed to accommodate Acremonium nepalense, Gliocladium cibotii and the new species B. macroclavatum and B. terrestre. Brunneochlamydosporium nepalense and B. macroclavatum were represented in our study with seven and one isolate respectively. According to Prenafeta-Boldú et al. (2014), the former species seems to be a common soil-borne fungus in the Netherlands, being present in Gelderland, North and South Holland and Utrecht provinces, while B. macroclavatum was isolated from a fern, and the flowering plant, Aphelandra sp. (Acanthaceae), but not from soil (Giraldo and Crous 2019).
Furcasterigmium furcatum was established based on Acremonium furcatum, and it was represented in our study with four isolates collected from North Brabant, South Holland and Utrecht provinces. Like B. nepalense, this species was recovered from soil in the Netherlands, but is also present in soils from France, Germany and Italy (Gams 1971; Giraldo and Crous 2019).
Two soil samples of Friesland and Gederland provinces revealed three isolates of Lectera that turned out to represent the new species, L. nordwiniana (Crous et al. 2018). Although most of the species in Lectera are plant pathogens, including L. colletotrichoides and L. capsici (Crous et al. 2017a), the former species is commonly isolated from soil and plant litter (Cannon et al. 2012). Another species recently described from soil is L. humicola from Brazil (Giraldo and Crous 2019).
In this study, Gibellulopsis and Plectosphaerella were the dominant genera, with G. nigrescens and Pl. plurivora as the most common species from each respective genus. In some studies of fungal biodiversity in soil, Gibellulopsis and Plectosphaerella are occasionally recovered (Hujslová et al. 2010; Duran et al. 2019; Wentzel et al. 2019). According to the data shown by Giraldo and Crous (2019), Gibellulopsis spp. inhabit soils around the world, with G. nigrescens recovered from Israel, the Netherlands, New Zealand and the UK. Species of Plectosphaerella are more common on plants than in soil, except for Pl. plurivora, which is present in soils of different countries in Northern Europe (Belgium, the Netherlands and Germany), and Pl. humicola and Pl. oligotrophica from Democratic Republic of the Congo, Brazil and China, respectively (Liu et al. 2013; Giraldo and Crous 2019). In our study, a single isolate (JW 83023) was identified as Pl. pauciseptata, known thus far from Citrullus lanatus, Cucumis melo, C. sativus and Solanum esculetum in Italy (Carlucci et al. 2012). Therefore, this is the first time that this species has been recovered from soil.
Other studies have described G. nigrescens from soil samples in Korea (Nguyen et al. 2018) and China (Wu et al. 2013), where it was also shown to be the cause of wilt of sugar beet (Zhou et al. 2017) and alfalfa (Hu et al. 2011). In Europe, this species and Plectosphaerella spp. have been found in saline and acidic soils in the Czech Republic (Hujslová et al. 2010), and inhabiting in good quality soils (as Gleyic Chernozem, Fluvic Cambisol, Cambic Leptosol) in Poland (Grządziel and Gałązka 2019). In a survey about microfungal community in cultivated soils, Pl. cucumerina and A. luteoalbus (as Verticillium tenerum) were found in low proportions in soils from different locations of the Eskişehir province in Turkey (Demirel et al. 2005). In Argentina, Pl. plurivora was found in rhizosphere soil, showing an antagonistic effect against eggs of the plant-parasitic nematode Nacobbus aberrans (Sosa et al. 2018), and Plectosphaerella sp. was recovered from farmland in the Tibetan Plateau (Li et al. 2012).
The studies based in Next Generation Sequence generally show a higher diversity of soil fungi compared with those based on culture-dependent methods (Tedersoo et al. 2014, 2017; Wardle and Lindahl 2014). The latter approach usually overlooks the true diversity, mainly because of the low percentage of fungi growing on artificial media. However, through this Citizen science project, we have explored the fungal diversity of Dutch soils, which has revealed a rich species diversity. Some of the taxa obtained represent new species, genera and even families, which have been described here or in other publications (Crous et al. 2017b, 2018; Groenewald et al. 2018), while others are still awaiting description (unpubl. data). As a result of this project, around 3000 fungal isolates were obtained from 293 soil samples. Among them, 386 isolates were yeast fungi which were treated by Groenewald et al. (2018), who identified 67 species distributed over 40 genera (including basidiomycetous and ascomycetous) including six new species in the genera Hanseniaspora, Ogataea, Pichia, Saccharomycopsis, Trichomonascus and Zygoascus. Regarding filamentous fungi, new taxa have been described in different and unrelated genera, including Acaulium, Collariella, Conioscypha, Fusarium, Fusicolla, Gamsia, Gibellulopsis, Lasionectria, Lectera, Leptodiscella, Parasarocladium, Phaeoisaria, Sarocladium, Striaticonidium, Talaromyces, Umbelopsis, Vandijckella (incl. Vandijckella gen. nov. and Vandijckellaceae fam.nov.) and Verhulstia trisororum (incl. Verhulstia gen. nov.). These results support previous findings concluding that soil contains numerous undescribed fungal taxa (Hujslová et al. 2010; Tedersoo et al. 2014, 2017; Wentzel et al. 2019) and highlights the importance to continue with these kinds of studies in order to generate isolates that can be available in public databases and can be freely used by other researchers.
References
Anderson JPE, Domsch KH (1978) A physiological method for the quantitative measurement of microbial biomass in soils. Soil Biol Biochem 10:215–221
Antignani V, Gams W, Marziano F (2008) Plectosporium delsorboi nov. sp., a pathogen of Curcuma, Zingiberaceae. Nova Hedwigia 86:209–214
Bills GF, Christensen M, Powell M et al (2004) Saprobic soil fungi. In: Mueller GM, Bills GF, Foster MS (eds) Biodiversity of fungi. Elsevier, London, pp 271–302
Blackwell M (2011) The fungi: 1, 2, 3... 5.1 million species? Am J Bot 98:426–438
Cannon P, Buddie AG, Bridge PD et al (2012) Lectera, a new genus of the Plectosphaerellaceae for the legume pathogen Volutella colletotrichoides. MycoKeys 3:23–36
Carlucci A, Raimondo ML, Santos J et al (2012) Plectosphaerella species associated with root and collar rots of horticultural crops in southern Italy. Persoonia 28:34–48
Crous PW, Gams W, Stalpers JA et al (2004) MycoBank: an online initiative to launch mycology into the 21st century. Stud Mycol 50:19–22
Crous PW, Wingfield MJ, Guarro J et al (2015) Fungal planet description sheets: 625–715. Persoonia 34:320–370
Crous PW, Wingfield MJ, Burgess TI et al (2017a) Fungal planet description sheets: 558–624. Persoonia 38:240–384
Crous PW, Wingfield MJ, Burgess TI et al (2017b) Fungal planet description sheets: 625–715. Persoonia 39:270–467
Crous PW, Luangsa-ard JJ, Wingfield MJ et al (2018) Fungal planet description sheets: 785–867. Persoonia 41:238–417
Crous PW, Verkley GJM, Groenewald JZ et al (2019) Fungal biodiversity. Westerdijk Laboratory Manual Series 1. Westerdijk Fungal Biodiversity Institute, The Netherlands
Demirel R, Ilhan S, Asan A et al (2005) Microfungi in cultivated fields in Eskişehir provience (Turkey). J Basic Microbiol 45:279–293
Domsch KH, Gams W, Anderson TH (2007) Compendium of soil fungi, 2nd edn. IHW Verlag, Germany
Duc PM, Hatai K, Kurata O et al (2009) Fungal infection of mantis shrimp (Oratosquilla oratoria) caused by two anamorphic fungi found in Japan. Mycopathologia 167:229–247
Duran P, Barra PJ, Jorquera MA et al (2019) Occurrence of soil fungi in Antarctic pristine environments. Front Bioeng Biotechnol 7:28
Gams W (1971) Cephalosporium-artige Schimmelpilze (Hyphomycetes). Gustav Fischer Verlag, Stuttgart
Giraldo A, Crous PW (2019) Inside Plectosphaerellaceae. Stud Mycol 92:227–286
Giraldo A, Gené J, Sutton DA et al (2017) New acremonium-like species in the Bionectriaceae and Plectosphaerellaceae. Mycol Prog 16:349–368
Groenewald M, Lombard L, de Vries M et al (2018) Diversity of yeast species from Dutch garden soil and the description of six novel Ascomycetes. FEMS Yeast Res. https://doi.org/10.1093/femsyr/foy076
Grum-Grzhimaylo AA, Debets AJM, van Diepeningen AD et al (2013) Sodiomyces alkalinus, a new holomorphic alkaliphilic ascomycete within the Plectosphaerellaceae. Persoonia 31:147–158
Grum-Grzhimaylo AA, Georgieva ML, Bondarenko SA et al (2016) On the diversity of fungi from soda soils. Fungal Divers 76:27–74
Grządziel J, Gałązka A (2019) Fungal biodiversity of the most common types of Polish soil in a long-term microplot experiment. Front Microbiol. https://doi.org/10.3389/fmicb.2019.00006.eCollection2019
Hu XP, Wang MX, Hu DF, Yang JR (2011) First report of wilt on alfalfa in China caused by Verticillium nigrescens. Plant Dis 95:1591
Hujslová M, Kubátová A, Chudíčková M et al (2010) Diversity of fungal communities in saline and acidic soils in the Soos National Natural Reserve, Czech Republic. Mycol Prog 9:1–15
Katoh K, Rozewicki J, Yamada KD (2017) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinformatics:1–7
Kirk PM, Cannon PF, David JC et al (2001) Dictionary of the fungi, 9th edn. CAB International, Wallingford
Li SL, Lin Q, Li XR et al (2012) Biodiversity of the oleaginous microorganisms in Tibetan plateau. Braz J Microbiol 43:627–634
Liu YJ, Whelen S, Hall BD (1999) Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 16:1799–1808
Liu TT, Hu DM, Liu F et al (2013) Polyphasic characterization of Plectosphaerella oligotrophica, a new oligotrophic species from China. Mycoscience 54:387–393
Mason-Gamer R, Kellogg E (1996) Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Syst Biol 45:524–545
Miller MA, Pfeiffer W, Schwartz T (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. In: Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the extreme to the campus and beyond: 1–8. Association for Computing Machinery, USA. https://doi.org/10.1109/GCE.2010.5676129
Nguyen TTT, Pangging M, Lee SH et al (2018) Four new records of ascomycete species from Korea. Mycobiology 64:328–340
Prenafeta-Boldú FX, Summerbell RC, De Boer W et al (2014) Biodiversity and ecology of soil fungi in a primary succession of a temperate coastal dune system. Nova Hedwigia 99:347–372
Rayner RW (1970) A mycological colour chart. Commonwealth Mycological Institute and British Mycological Society, Kew
Réblová M, Gams W, Seifert KA (2011) Monilochaetes and allied genera of the Glomerellales, and a reconsideration of families in the Microascales. Stud Mycol 68:163–191
Rehner SA, Buckley E (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97:84–98
Sosa AL, Rosso LC, Salusso FA et al (2018) Screening and identification of horticultural soil fungi for their evaluation against the plant parasitic nematode Nacobbus aberrans. World J Microbiol Biotechnol 34:63
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1213
Su L, Deng H, Niu YC (2017) Phylogenetic analysis of Plectosphaerella species based on multi-locus DNA sequences and description of P. sinensis sp. nov. Mycol Prog 16:823–829
Summerbell RC, Gueidan C, Schroers HJ et al (2011) Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud Mycol 68:139–162
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tedersoo L, Bahram M, Põlme S et al (2014) Global diversity and geography of soil fungi. Science 346:1256688–1–1256688-10
Tedersoo L, Bahram M, Puusepp R et al (2017) Novel soil-inhabiting clades fill gaps in the fungal tree of life. Microbiome. https://doi.org/10.1186/s40168-017-0259-5
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246
Vilgalys R, Sun BL (1994) Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proc Natl Acad Sci 91:4599–4603
Wardle DA, Lindahl BD (2014) Disentangling global soil fungal diversity. Science 346:1052–1053
Wentzel LCP, Inforsato FJ, Montoya QV et al (2019) Fungi from Admiralty Bay (King George Island, Antarctica) soils and marine sediments. Microb Ecol 77:12–24
White TJ, Burns T, Lee S et al (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 282–287
Wiens JJ (1998) Testing phylogenetic methods with tree congruence: phylogenetic analysis of polymorphic morphological characters in phrynosomatid lizards. Syst Biol 47:427–444
Wu YM, Xu JJ, Wang HF et al (2013) Geosmithia tibetensis sp. nov. and new Gibellulopsis and Scopulariopsis records from Qinghai-Tibet. Mycotaxon 125:59–64
Zare R, Gams W, Starink-Willemse M et al (2007) Gibellulopsis, a suitable genus for Verticillium nigrescens, and Musicillium, a new genus for V. theobromae. Nova Hedwigia 85:463–489
Zhou Y, Zhao ZQ, Guo QY et al (2017) First report of wilt of sugar beet caused by Gibellulopsis nigrescens in the Xinjiang region of China. Plant Dis 101:1318
Acknowledgements
We are grateful to all the children, parents and teachers that enthusiastically participated in the collecting and submission of samples to the Westerdijk Institute. We are thankful to the Utrecht University Museum and KNAW for funding and promoting the project among the Dutch primary schools; to the Utrecht University for coordinating the initiative ‘meet the professor’; to the staff from the Westerdijk Institute: Manon Verweij, Karin Schagen and Mariëtte Oosterwegel for promoting the project and establishing communication with the collectors and schools. We also thank the CBS collection staff, especially Trix Merkx and Arien van Iperen for depositing the isolates and specimens in the culture collection and fungarium. Additional thanks are extended to the MycoBank curator, Konstanze Bench, for her suggestions and for checking the spelling of the epithets of the new species, and Duong Vu for her assistance with the database.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editorial Responsibility: Marc Stadler
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Giraldo, A., Hernández-Restrepo, M. & Crous, P.W. New plectosphaerellaceous species from Dutch garden soil. Mycol Progress 18, 1135–1154 (2019). https://doi.org/10.1007/s11557-019-01511-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-019-01511-4