Abstract
During a survey of Xylariales in northern Thailand, several specimens with affinities to the genus Daldinia were found and examined for morphological characters, secondary metabolites, and molecular phylogenetic traits. Aside from morphological and chemotaxonomic studies, a multi-locus phylogenetic analysis using internal transcribed spacers regions (ITS) and the large subunit (LSU) of the ribosomal DNA, the second largest subunit of the RNA polymerase (RPB2), and beta-tubulin (TUB2) genes was performed. Among the specimens was a new species and a new record of a species that had previously never been sequenced and studied for its anamorphic morphology. This species, previously described by Ju and Rogers as Hypoxylon kretzschmarioides based on a single record from Indonesia, showed secondary metabolite profiles reminiscent of those of the genus Daldinia and even clustered in the latter genus in the phylogenetic tree. Therefore, it is transferred to Daldinia as D. kretzschmarioides comb. nov. A second new species, D. subvernicosa sp. nov., was found to have a close relationship with D. vernicosa based on morphological and molecular evidence, but differs from D. vernicosa by long-stipitate asci with mostly subglobose ascospores, and the basal ascospores are often elongated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Daldinia was described by Cesati and De Notaris (1863) and belongs to the Hypoxylaceae (Xylariales), since the recent rearrangement of the families of stromatic Xylariales by Wendt et al. (2018). The Hypoxylaceae is one of the largest families in this order and both the family and the genus Daldinia have been studied exhaustively for secondary metabolite production (Helaly et al. 2018). Daldinia was traditionally separated from Hypoxylon based on the presence of internal concentric zones in their stromata (Ju et al. 1997). However, D. placentiformis (Berk. and M.A. Curtis) Theiss. (1909) has for long been included in the genus Hypoxylon, to which it had belonged until Hsieh et al. (2005) provided evidence from molecular phylogenetic data that its affinities are indeed with Daldinia. This was later confirmed in the chemotaxonomic study by Bitzer et al. (2008). In the world monograph by Stadler et al. (2014), the genus was segregated by using a combination of morphological, chemotaxonomic, and molecular phylogenetic characters. While only ITS data had been used in the latter study, Wendt et al. (2018) have included several species and demonstrated by using a multi-locus phylogeny that Daldinia and allied species indeed represent an independent lineage in the Hypoxylaceae that is different from Hypoxylon as well as from the genus Pyrenopolyporus, which was resurrected and amended to accommodate some species with superficial similarities to D. placentiformis. A more comprehensive overview by Daranagama et al. (2018) includes a backbone phylogeny of important taxa in the Xylariaceae and other families of stromatic Xylariales and provided updated descriptions and illustrations for all taxa, thus serving as valuable reference. In Thailand and other Asian countries, the genus still needs more research.
During our ongoing surveys of Xylariales in northern Thailand, we have encountered two interesting Daldinia species, of which one represents a new taxon and the other shows affinities to another species that has so far only been found once in Indonesia. The present study is dedicated to the presentation of their morphological and chemotaxonomic features and their phylogenetic placement.
Materials and methods
Morphological characterization
Measurements of morphological characters, such as size and shapes of stromata, perithecia, asci, and ascospores, were examined according to Stadler et al. (2014). The cultures of the specimens were obtained from multiple spore isolation following Sir et al. (2016a). Preliminary classification was done by examining the conidiogenous cells and conidiophore branching pattern of the asexual morph according to Ju and Rogers (1996). Furthermore, the stromatal color, KOH extractable pigment, and cultures are recorded according to Rayner (1970). The cultures and the material vouchers were deposited in Thailand Bioresource Research Center (TBRC) and BIOTEC Bangkok Herbarium (BBH), respectively. Scanning electron microscopy (SEM) was carried out using a conventional procedure described by Kuhnert et al. (2017).
HPLC profiling
For chemotaxonomic studies, the natural products were extracted using the method by Yuyama et al. (2018), using high performance liquid chromatography coupled with diode array and electrospray mass spectrometric detection (HPLC/DAD-ESIMS). The instrumental settings and conditions were as described in Kuhnert et al. (2017).
DNA extraction, PCR, and phylogenetic analyses
The mycelium was extracted using cetyltrimethyl ammonium bromide (CTAB) following the method by Mackill and Bonman (1995). Four DNA loci including internal transcript spacer regions (ITS); large subunit of the rDNA (LSU); RNA polymerase II (RPB2); and beta tubulin (TUB2) were amplified by PCR, following the standard primers introduced by White et al. (1990; ITS1, ITS4, and ITS5), Vilgalys and Hester (1990; LR7 and LROR), Liu et al. (1999: RPB2–5F and 7Cr), and O’Donnell and Cigelnik (1997; T1 and T22) following the protocols of Otto et al. (2016) and Wendt et al. (2018). DNA sequences were checked and assembled using BioEdit v. 7.2.5 (Hall 2013). The new sequences were submitted to GenBank (Table 1). The molecular analyses were done following Wendt et al. (2018). All sequences were then aligned using MUSCLE (Edgar 2004) and alignments were refined by direct examination. Multiple sequence alignments were analyzed with the closely matched sequences obtained from GenBank (Table 1). Sequences were analyzed using maximum parsimony (MP), maximum likelihood (ML), and Bayesian algorithm. Maximum parsimony analysis was performed in PAUP*4.0b10 Swofford (2002). The most parsimonious trees (MPTs) were obtained from the heuristic searches: 100 replicates of random stepwise addition of sequence, branch-swapping algorithm, tree-bisection-reconnection (TBR), and equal weight characters. Maximum parsimony bootstrap supports were estimated by 1000 replicates (stepwise addition of sequence, 10 replicates of random addition of taxa, TBR branching-swapping algorithm). Most parsimonious tree length, consistency index (CI), retention index (RI), relative consistency index (RC), and homoplasy index (HI) were estimated. The maximum likelihood and bootstrap analyses were conducted through the CIPRES web portal (Miller et al. 2010) using RAxML 8.2.4 (Stamatakis 2014) with the BFGS method to optimize GTR rate parameters. Finally, Bayesian posterior probabilities of the branches were performed using MrBayes 3.0B4 (Huelsenbeck and Ronquist 2001) with the best-fit model (GTR+I+G) selected by AIC in Mr Modeltest 2.2 (Nylander 2004) that was tested with hierarchical likelihood ratios (hLRTs). Three million generations were run in four Markov chains and sampled every 100 generations with a burn in value set at 3000 sampled trees
Results and discussion
Taxonomy
Daldinia kretzschmarioides (Y.M. Ju & J.D. Rogers) Srikitikulchai, Wongkanoun, M. Stadler & Luangsa–ard, comb. nov. Fig. 1.
Daldinia kretzschmarioides (BBH 42281). a–b Stromata in wood; c stromatal surface with ostioles; d cross section of stroma showing perithecia and the tissue below the perithecial layer (white arrow); e perithecia (white arrow); f ascus; g apical apparatus bluing in Melzer’s reagent (black arrow); h ascospore; i ascospore showing germ slit; j ascospore in KOH showing dehiscent perispore (black arrow); k pigments in 10% KOH. Scale is indicated by bars (a 20 mm. b 10 mm. d 5 mm. e 1 mm. f 10 μm, g–j 5 μm)
MB829270
Basionym: Hypoxylon kretzschmarioides Y.M. Ju & J.D. Rogers, Mycol. Mem. 20: 139 (1996)
Epitype (designated here): Thailand: Chiang Mai Province, Ban Hua Thung community forest, 19.42044′ N, 98.97140′ E, on dead angiosperm in the forest, 3 November 2016, P. Srikitikulchai, S. Wongkanoun, BBH 42276 (MBT383621)
Ex-epitype strain: TBRC 8875 (BBC); DNA sequences of ex-epitype strain: MH938531 (ITS), MH938540 (LSU), MK165425 (RBP2), MK165416 (TUB2)
Teleomorph.Stroma superficial, small to widely effused, pulvinate or peltate, the base broadly attached to the substrate, conspicuous or inconspicuous perithecial mounds, 25–29 mm long × 9.45–13 (27) mm broad × 2–3 mm thick; surface mouse gray (118) to pale mouse gray (117) brownish yellow or red-orange granules forming a thin crust above perithecia, with 10% KOH producing dark vinaceous (82) extractable pigments, the tissue between perithecia gray or blackish brown, the tissue below perithecia layer gray, 1.2–2.4 mm thick. Perithecia monostichous, lanceolate, 0.14–0.28 mm broad × 1.40–1.42 mm high; ostioles black, umbilicate. Asci cylindrical, spore bearing part 60–63 μm long × 8 μm; apical apparatus bluing in Melzer’s reagent, discoid, 0.5–1 × 2.5–3 μm. Ascospores dark brown to blackish brown, unicellular, ellipsoid, (4) 5–6 × 13–15 (16) μm (mean = 5.13 × 13.83 μm, n = 30) with straight to slightly oblique germ slit much less than spore length on convex size, perispore dehiscent in 10% KOH, smooth.
Anamorph in culture. Conidiophores with virgariella-like to (much more frequently) nodulisporium-like branching patterns as defined in Ju and Rogers (1996). Main axis hyaline and cell walls rough or smooth dark brown to blackish brown. Conidiogenous cells cylindrical, hyaline, finely roughened, 10–17 × 3–4 μm. Conidia hyaline, smooth, ellipsoid 5−7 × 3−4 μm.
Culture characteristics. Colonies on OA reaching the edge of a 9 cm Petri dish in 1 week, at first whitish becoming velvety to felty, azonate with entire margin, grayish yellow–green (68), olivaceous (48) and dark herbage green (69) to dull green (70) after 2 weeks incubation (Fig. 2e). Colonies on YMGA covering Petri dish in 1 week at first whitish becoming smoke gray, dark herbage green (69), and dull green (70) velvety to felty, azonate with entire margin.
Daldinia kretzschmarioides (TBRC 8875). a asexual morph showing conidiophores with virgariella-like to nodulisporium-like branching patterns; b nodulisporium-like branching patterns, conidiogenous cells (arrows); c conidiogenous cell (arrow); d conidia; e culture on OA medium after 2 weeks. Scale is indicated by bars (a–b 20 μm. c–d 10 μm. e 2 cm)
Secondary metabolites. BNT, Cytochalasins
Notes. The specimen showed very similar characteristics to the holotype of the monotypic species Hypoxylon kretzschmarioides, which originates from Indonesia and has never been cultured or subjected to DNA sequencing. As already mentioned by Wendt et al. (2018), Ju et al. (1997) have described in the protologue that the perispore of the ascospores of this specimen was indehiscent, but a re-examination of the type specimen in NY (J. Fournier and M.S., unpublished) had revealed that the perispore is actually dehiscent. All other salient morphological characters of the Thai specimen that we propose as epitype of H. kretzschmarioides are in agreement with the holotype. Therefore, we regard the current specimen as conspecific to H. kretzschmarioides. Since the results of the molecular phylogeny leave no doubt that the phylogenetic affinities of the fungus are with the genus Daldinia, we have moved H. kretzschmarioides to the latter genus.
There are two other Daldinia species with similar stromatal morphology, lacking internal concentric zones: Daldinia kretzschmarioides is morphologically similar to the circumtropically distributed D. placentiformis but differs in its ascospore size range as well as in having olivaceous stromatal pigments, owing to the presence of daldinone A as predominant stromatal metabolite. The Argentine species, Daldinia korfii (cf. Sir et al. 2016b) is also similar but differs in its ascospore size range. HPLC profiling showed that both the holotype and the selected epitype specimen contained BNT and cytochalasins (Table 2). The BNT peak was more prominent in the epitype, which explains the stronger purple color as compared to the holotype specimen in NY, which had been collected several years previously. The major cytochalasins in the stromata of the Thai specimens were recently identified as the new phenochalasins C and D (Figs. 3 and 4) and found to exhibit significant anti-biofilm effects in Staphylococcus aureus (Yuyama et al. 2018).
Daldinia subvernicosa Srikitikulchai, Wongkanoun, M. Stadler & Luangsa–ard, sp. nov. Fig. 5.
Daldinia subvernicosa (BBH 42276). a stromatal habit; b stromatal surface with ostioles; c perithecia; d cross section of stroma showing alternating zones; e concentric zones; f ascus; g ascospore in distilled water; h–i ascospore by SEM; j colony on OA medium for 1 week. Scale is indicated by bars (e 0.5 mm. f 10 μm. g–i 5 μm. j 2 mm)
MB828032
Etymology. In reference to the morphological similarities to Daldinia vernicosa
Holotype: Thailand: Chiang Mai Province, Ban Hua Thung community forest, 19.42044′ N, 98.97140′ E, on dead angiosperm in the forest, 3 November 2016, P. Srikitikulchai and S. Wongkanoun, BBH 42281
Ex-holotype strain: TBRC 8877 (BBC). DNA sequences of ex-holotype strain: MH938533 (ITS), MH938542 (LSU), MK165430 (RPB2), MK165421 (TUB2)
Teleomorph.Stroma hemispherical to depressed-spherical, widely attached to the substrate, very rarely substipitate, smooth or with inconspicuous perithecial outline, 2.90–5 cm × 1.68–3.40 cm; surface fuscous black (104), with 10% KOH extractable pigments mouse gray (118), dark brown to dark black immediately beneath the surface; tissue between perithecia blackish brown, woody; tissue below the perithecia layer composed of alternating zones, darker zones blackish brown, 0.1 mm thick, lighter zones, white, 1 mm thick. Perithecia subglobose 1 mm diam; ostioles umbilicate, lower than the stromatal surface. Asci unitunicate, cylindrical, 220–236 long, with long stipe, 135–143 μm, the spore bearing part 85–93 × 13–15 μm, 8-spored, without visible apical apparatus, not bluing in Melzer’s reagent. Ascospores unicellular, dark brown to blackish brown, (5)–8–10 × 12–15 μm (mean = 9.25 × 13.44 μm, n = 100), rectangular, subglobose, often oriented transverse to the ascal axis, the basal ascospore often ellipsoid, oblong to elongate, with conspicuous germ slit spore length, without dehiscing layer in 10% KOH.
Culture characteristics. Colonies on OA covering the edge of 9 cm. Petri dish in 7 days, at first whitish, floccose, azonate, becoming smoke gray (105) and isabelline (65) (Fig. 3j). After 3 weeks incubation, the fungus produced synnemata on the agar medium but did not sporulate. Colonies on YMGA covering the edge of a 9 cm Petri dish in 6–7 days, at first whitish, becoming smoke gray (105), velvety to felty, azonate with entire margin.
Secondary metabolite. BNT (binaphthalene tetrol).
Notes. The closest relative of the new species is clearly D. vernicosa, which has eventually been regarded as cosmopolitan by Child (1932) and Ju et al. (1997, as D. fissa) but was only encountered in the more comprehensive study by Stadler et al. (2014) among the specimens originating from the temperate Northern hemisphere. A peculiar feature of D. vernicosa is that this species readily produces not only a very characteristic virgariella-like anamorph in culture but often forms stromata, in particular on Oatmeal agar (Ju et al. 1999; Stadler et al. 2014). Daldinia loculata (Lév.) Sacc. is closely related with D. subvernicosa but differs in stromatal and ascospore morphology. Daldinia loculatoides Wollweber & M. Stadler also has affinities to the new taxon, but has more regular ascospores and brown internal concentric zones. In addition, the species is only known from the temperate Northern hemisphere. Daldinia singularis Y.M. Ju, Lar.N. Vassiljeva & J.D. Rogers also is lacking a well-developed apical apparatus but differs by having smaller ascospores and a different anamorphic structure. Daldinia bakeri Lloyd (1919) is highly similar to D. subvernicosa in the shape and color of stroma while the ascospore length but has much larger ascospores than D. subvernicosa. The molecular phylogeny (Fig. 6) also confirmed the status of D. subvernicosa as a new species.
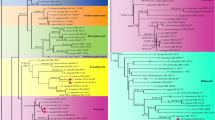
Phylogeny of the Hypoxylaceae. The phylogenetic relationships are depicted as RA × ML tree was generated base on genetic multiples loci alignment of ribosomal (ITS and LSU) and proteinogenic (TUB2 and RPB2) sequence information. In maximum parsimony analysis, a CI of 0.372, a RI of 0.572, and a HI of 0.628 yielded only one parsimony tree with a length of 8802 changes. The phylogenetic relationships inferred from RA × ML had a likelihood of − 43934.190 and likelihood of the Bayesian tree was − 43244.290. Support values were calculated via maximum parsimony (MP), maximum likelihood (ML), and Bayesian analysis; and are indicated above (MP/ML) and below (B) the respective branches, if the bootstrap support (BS) values exceeded 50% from 1000 replicates or the posterior probability (PP) value from 3,000,000 MCMC chains (sampling frequency 1000, 10% burn-in) was 0.95 or higher. Branches of significant support (BS ≥ 95% and PP ≥ 0.98) are thickened
Molecular phylogeny
Twenty-seven new sequences were generated from the amplification of ITS, LSU, RPB2, and TUB2 regions (Table 1). These gene regions were selected to clarify the phylogenetic relationships of Daldinia and how they differ from other species and genera in the Hypoxylaceae. PCR amplifications yielded approx. 500 bp of ITS rDNA, 1000 bp of the LSU rDNA, approx. 800 bp of the RPB2, and approx. 1000 bp of the TUB2 region that were selected to clarify the phylogenetic relationships of several genera belonging to the Hypoxylaceae. The phylogenetic relationships were estimated using maximum parsimony (MP) and maximum likelihood (ML) analyses. The dataset of the multi-loci DNA sequences including 51 taxa in the Hypoxylaceae comprising 5 taxa in Annulohypoxylon, 23 taxa of Daldinia, 11 taxa of Hypoxylon, 2 taxa of Hypomontagnella, 3 taxa of Jackrogersella, and 5 taxa of Pyrenopolyporus with Graphostoma platystomum and Xylaria hypoxylon used as the out groups. The combined dataset consists of 4451 characters, of which 2578 were constant, 1380 parsimony informative, and 493 un-informative. The best tree generated through maximum parsimony analysis yielded only one most parsimonious tree. The molecular analyses revealed that DNA sequences are placed in the Hypoxylaceae. The phylogenetic tree including 4 major clades comprising the top clade is Daldinia clade and the Pyrenopolyporus clade, Hypomontagnella, Annulohypoxylon, Jackrogersella, and the lower clade as Hypoxylon, respectively (Fig. 6). The upper clade forms a monophyletic clade consisting of D. eschscholtzii, D. placentiformis, and D. theissenii placed in clade D1 as a cryptic clade. However, we need more samples of D. eschscholtzii to fill the taxonomic database. Clade D2 formed a monophyletic group with high support consisting of D. bambusicola and D. caldariorum. Our two samples (TBRC 8878, TBRC 8879) were placed in this clade with closest affinities to D. bambusicola, forming a sister clade to clade D1. Besides, the clade D3 forms a distinct clade within the genus Daldinia with strong statistical support (100% BSMP, 100% BSML, and 1.00 BYPP), with D1 and D2 as sister clades and consists of D. korfii and D. kretzschmarioides comb. nov. In agreement with the morphological characteristics, and the two taxa are separated with high statistical support. Clade D4, consisting of D. andina, D. concentrica, D. dennisii, D. loculatoides, D. macaronesica, and D. steglichii, also formed a sister clade with clade D5. The clade D5 was comprised of two subclades, one of which included D. petriniae and D. pyrenaica, while the other contained D. subvernicosa sp. nov. and D. vernicosa. The clade PY consisted of Pyrenopolyporus species and in agreement with Wendt et al. (2018) was a sister clade to D4. The clade HY, contained representatives of the recently erected genus Hypomontagnella (Lambert et al. 2019) with H. monticulosa and H. submonticulosa. The clade A included species of Annulohypoxylon and clade J, the members of Jackrogersella, appeared as a sister clade with clade A. The clade H comprised the species of Hypoxylon. The latter findings are in accordance with Wendt et al. (2018).
References
Bitzer J, Læssøe T, Fournier J, Kummer V, Decock C, Tichy HV, Piepenbring M, Peršoh D, Stadler M (2008) Affinities of Phylacia and the daldinoid Xylariaceae, inferred from chemotypes of cultures and ribosomal DNA sequences. Mycol Res 112:251–270
Cesati V, De Notaris G (1863). Schema di classificazione degli Sferiacei italici aschigeri più o meno appartenenti al genere Sphaeria nelľ antico significato attribuitoglide Persoon. Commentario della Societa Crittogamologica Italiana 1(4):177–420.
Child M (1932) The genus Daldinia. Ann Mo Bot Gard 19:429–496
Daranagama DA, Camporesi E, Tian Q, Liu X, Chamyuang S, Stadler M, Hyde KD (2015) Anthostomella is polyphyletic comprising several genera in Xylariaceae. Fungal Divers 73:203–238
Daranagama DA, Hyde KD, Sir EB, Thambugala KM, Tian Q, Samarakoon MC, McKenzie EHC, Jayasiri SC, Tibpromma S, Bhat JD, Liu X, Stadler M (2018) Towards a natural classification and backbone tree for Graphostromataceae, Hypoxylaceae, Lopadostomataceae and Xylariaceae. Fungal Divers 88:1–165
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Hall TA (2013) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Helaly SE, Thongbai B, Stadler M (2018) Diversity of biologically active secondary metabolites from endophytic and saprotrophic fungi of the ascomycete order Xylariales. Nat Prod Rep 35:992–1014
Hsieh HM, Ju YM, Rogers JD (2005) Molecular phylogeny of Hypoxylon and closely related genera. Mycologia 97:844–865
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Johannesson H, Laessøe T, Stenlid J (2000) Molecular and morphological investigation of the genus Daldinia in Northern Europe. Mycol Res 104:275–280
Ju YM, Rogers JD (1996) A revision of the genus Hypoxylon. Mycologia memoir no.° 20. APS Press, St. Paul 365 pp
Ju YM, Rogers JD, San Martín F (1997) A revision of the genus Daldinia. Mycotaxon 61:243–293
Ju YM, Vasilyeva L, Rogers JD (1999) Daldinia singularis sp. nov. from Eastern Russia and notes on some other taxa. Mycotaxon 71:405–412
Koukol O, Kelnarová I, Černý K (2015) Recent observations of sooty bark disease of sycamore maple in Prague (Czech Republic) and the phylogenetic placement of Cryptostroma corticale. For Pathol 45:21–27
Kuhnert E, Fournier J, Peršoh D, Luangsa-ard JJ, Stadler M (2014) New Hypoxylon species from Martinique and new evidence on the molecular phylogeny of Hypoxylon based on ITS rDNA and β-tubulin data. Fungal Divers 64:181–203
Kuhnert E, Sir EB, Lambert C, Hyde KD, Hladki AI, Romero AI, Rohde M, Stadler M (2017) Phylogenetic and chemotaxonomic resolution of the genus Annulohypoxylon (Xylariaceae) including four new species. Fungal Divers 85:1–43
Lambert C, Wendt L, Hladki AI, Stadler M, Sir EB (2019) Hypomontagnella (Hypoxylaceae): a new genus segregated from Hypoxylon by a polyphasic taxonomic approach. Mycol Prog. https://doi.org/10.1007/s11557-018-1452-z
Liu YL, Whelen S, Hall BD (1999) Phylogenetic relationships among ascomycetes: evidence from and RNA polymerase II subunit. Mol Biol Evol 16:1799–1808
Lloyd CG (1919) The genus Daldinia. Mycol Writ 5:23–26
Mackill DJ, Bonman JM (1995) Classifying japonica rice cultivars with RAPD markers. Crop Sci 35:889–894
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. Gateway computing environments workshop (GCE), IEEE, San Diego, Supercomputer Center, La Jolla, CA, USA, Nov 14, pp 1–8
Nylander JAA (2004) MrModeltest v. 2.0. Evolutionary biology centre Uppsala University (Program distributed by the author)
O’Donnell K, Cigelnik E (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 7:103–116
Otto A, Laub A, Wendt L, Porzel A, Schmidt J, Palfner G, Becerra J, Krüger D, Stadler M, Wessjohann L, Westermann B, Arnold N (2016) Chilenopeptins A and B, peptaibols from the Chilean Sepedonium aff. chalcipori KSH 883. J Nat Prod 79:929–938
Rayner RW (1970) A mycological colour chart. Commonwealth Mycological Institute, Kew and British Mycological Society
Sir EB, Kuhnert E, Lambert C, Hladki AI, Romero AI, Stadler M (2016a) New species and reports of Hypoxylon from Argentina recognized by a polyphasic approach. Mycol Prog 15:42
Sir EB, Lambert C, Wendt L, Hladki AI, Romero AI, Stadler M (2016b) A new species of Daldinia (Xylariaceae) from the Argentine subtropical montane forest. Mycosphere 7(5):596–614
Stadler M, Kuhnert E, Peršoh D, Fournier J (2013) The Xylariaceae as model example for a unified nomenclature following the “One Fungus-One Name” (1F1N) Concept. Mycology 4:5–21
Stadler M, Læssøe T, Fournier J, Decock C, Schmieschek B, Tichy HV, Peršoh D (2014) A polyphasic taxonomy of Daldinia (Xylariaceae). Stud Mycol 77:1–143
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Swofford DL (2002) PAUP*4.0b10: phylogenetic analysis using parsimony (*and other methods). Sinauer, Sunderland. https://doi.org/10.1111/j.0014-3820.2002.tb00191.x
Theissen F (1909) Xylariaceae austro-brasilienses. Zweiter Teil. Ann Mycol 7:1–18
Triebel D, Peršoh D, Wollweber H, Stadler M (2005) Phylogenetic relationships among Daldinia, Entonaema and Hypoxylon as inferred from ITS nrDNA sequences. Nova Hedw 80:25–43
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4239–4246
Wendt L, Sir EB, Kuhnert E, Wendt L, Sir EB, Kuhnert E, Heitkämper S, Lambert C, Hladki AI, Romero AI, Luangsa-ard JJ, Srikitikulchai P, Peršoh D, Stadler M (2018) Resurrection and emendation of the Hypoxylaceae, recognised from a multi-gene genealogy of the Xylariales. Mycol Prog 17:115–154
White TJ, Bruns L, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Chapter 38. In: Innis M, Gelfand D, Sninsky J, White T (eds) PCR Protocols: a Guide to Methods and Applications. Academic Press, Orlando, pp 315–322
Yuyama KT, Wendt L, Surup F, Kretz R, Chepkirui C, Wittstein K, Boonlarppradab C, Wongkanoun S, Luangsa-ard JJ, Stadler M, Abraham WR (2018) Cytochalasans act as inhibitors of biofilm formation of Staphylococcus aureus. Biomolecules 8:129
Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, Seifert KA, Rossman AY, Rogers JD, Kohlmeyer J, Volkmann-Kohlmeyer B, Sung GH (2006) An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 98:1076–1108
Acknowledgments
We acknowledge Dr. Mathias Müsken for SEM recordings and Dr. Satinee Suetrong for molecular analysis suggestion. Our warmest thanks also go to Ms. Jirawan Kumsao for sample collections in Ban Hua Thung community forest in northern Thailand.
Funding
This work was supported by the National Science and Technology Development Agency (NSTDA), Cluster and Management Program Office (CPMO) for the project “Identification of Xylariaceae by induction of fruiting bodies formation” grant number P-12-01878; the National Center for Genetic Engineering and Biotechnology for RI project “Surveys and Collection Invertebrate-Pathogenic Fungi and Xylariaceae on Forests Conservation of Thailand” grant number P-14-51240; the European Union’s Horizon 2020 research and innovation program (RISE) under the Marie Skłodowska-Curie grant agreement no. 645701, project acronym “GoMyTri”; lead beneficiaries JJL and MS.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Roland Kirschner
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Taxonomic novelties: Daldiniasubvernicosa Srikitikulchai, Wongkanoun, M. Stadler & Luangsa–ard, sp. nov.; D. kretzschmarioides (Y.M. Ju & J. D. Rogers) Srikitikulchai, Wongkanoun, M. Stadler&Luangsa–ard, comb. nov
Rights and permissions
OpenAccess This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wongkanoun, S., Wendt, L., Stadler, M. et al. A novel species and a new combination of Daldinia from Ban Hua Thung community forest in the northern part of Thailand. Mycol Progress 18, 553–564 (2019). https://doi.org/10.1007/s11557-019-01469-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-019-01469-3